Implementation of Discretisation and Correlation-based Feature Selection to Optimize Support Vector Machine in Diagnosis of Chronic Kidney Disease
DOI:
https://doi.org/10.12928/biste.v5i2.7548Keywords:
Support Vector Machine, Discretization, CFS, Chronic Kidney DiseaseAbstract
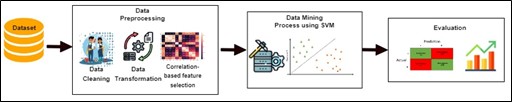
This study aims to improve the accuracy of the classification algorithm for diagnosing chronic kidney disease. There are several models of data mining. In classification, the Support Vector Machine (SVM) algorithm is widely used by researchers worldwide. The data used is a chronic kidney disease dataset taken from the UCI machine learning repository. This data consists of 25 attributes and 11 numeric data attributes, and 14 negative attributes. To call continuously, discrete data is used. Meanwhile, data is selected using Correlation-based Feature Selection (CFS) to reduce irrelevant and redundant data. The research results by applying discretization and feature selection based on correlation for classification in the SVM algorithm with 10-fold cross-validation show an increase in accuracy of 0.5%. The classification of the vector machine support algorithm in the diagnosis of chronic kidney disease produces an accuracy of 99.25%, and after applying discretization and correlation-based feature selection, produces an accuracy of 99.75%. Implementation of discretion and correlation-based feature selection to optimize support vector machine for diagnosis of chronic kidney disease has increased accuracy by 0.5%. The proposed method is feasible as a method of diagnosing chronic kidney disease.
References
M. S. Kukasvadiya and N. H. Divecha, “Analysis of data using data mining tool orange,” Int. J. Eng. Dev. Res., vol. 5, no. 2, pp. 1836–1840, 2017, https://www.ijedr.org/viewfull.php?&p_id=IJEDR1702288.
A. Kumar, P. Kumar, A. Srivastava, V. D. Ambeth Kumar, K. Vengatesan, and A. Singhal, “Comparative analysis of data mining techniques to predict heart disease for diabetic patients,” in International Conference on Advances in Computing and Data Sciences, pp. 507–518, 2020, https://doi.org/10.1007/978-981-15-6634-9_46.
S. A. Salloum, M. Al-Emran, A. A. Monem, and K. Shaalan, “Using text mining techniques for extracting information from research articles,” in Intelligent natural language processing: Trends and Applications, pp. 373–397, 2018, https://doi.org/10.1007/978-3-319-67056-0_18.
S. E. Bibri and J. Krogstie, “The big data deluge for transforming the knowledge of smart sustainable cities: A data mining framework for urban analytics,” in Proceedings of the 3rd International Conference on Smart City Applications, pp. 1–10, 2018, https://doi.org/10.1145/3286606.3286788.
R.-J. Kuo, T. C. Lin, F. E. Zulvia, and C. Y. Tsai, “A hybrid metaheuristic and kernel intuitionistic fuzzy c-means algorithm for cluster analysis,” Appl. Soft Comput., vol. 67, pp. 299–308, 2018, https://doi.org/10.1016/j.asoc.2018.02.039.
S. Feng, H. Zhou, and H. Dong, “Using deep neural network with small dataset to predict material defects,” Mater. Des., vol. 162, pp. 300–310, 2019, https://doi.org/10.1016/j.matdes.2018.11.060.
M. R. Hidayah, I. Akhlis, and E. Sugiharti, “Recognition number of the vehicle plate using Otsu method and K-nearest neighbour classification,” Sci. J. Informatics, vol. 4, no. 1, pp. 66–75, 2017, https://doi.org/10.15294/sji.v4i1.9503.
S. T. Ikram and A. K. Cherukuri, “Intrusion detection model using fusion of chi-square feature selection and multi class SVM,” J. King Saud Univ. Inf. Sci., vol. 29, no. 4, pp. 462–472, 2017, https://doi.org/10.1016/j.jksuci.2015.12.004.
J. Jumanto, M. A. Muslim, Y. Dasril, and T. Mustaqim, “Accuracy of Malaysia Public Response to Economic Factors During the Covid-19 Pandemic Using Vader and Random Forest,” J. Inf. Syst. Explor. Res., vol. 1, no. 1, pp. 49–70, 2023, https://doi.org/10.52465/joiser.v1i1.104.
H. A. Winarno, A. I. Poernama, I. Soesanti, and H. A. Nugroho, “Evaluation on EMG Electrode Reduction in Recognizing the Pattern of Hand Gesture by Using SVM Method,” J. Phys. Conf. Ser., vol. 1577, no. 1, 2020, https://doi.org/10.1088/1742-6596/1577/1/012044.
A. Toha, P. Purwono, and W. Gata, “Model Prediksi Kualitas Udara dengan Support Vector Machines dengan Optimasi Hyperparameter GridSearch CV”, Buletin Ilmiah Sarjana Teknik Elektro, vol. 4, no. 1, pp. 12–21, May 2022, https://doi.org/10.12928/biste.v4i1.6079.
Triwiyanto, O. Wahyunggoro, H. A. Nugroho, and Herianto, “Upper Limb Elbow Joint Angle Estimation Based on Electromyography Using Artificial Neural Network,” in 2018 12th South East Asian Technical University Consortium (SEATUC), pp. 1–6, 2018, https://doi.org/10.1109/SEATUC.2018.8788877.
D. A. Pisner and D. M. Schnyer, “Support vector machine,” in Machine learning, pp. 101–121, 2020, https://doi.org/10.1016/B978-0-12-815739-8.00006-7.
R. Rosita, D. A. A. Pertiwi, and O. G. Khoirunnisa, “Prediction of Hospital Intesive Patients Using Neural Network Algorithm,” J. Soft Comput. Explor., vol. 3, no. 1, pp. 8–11, 2022, https://doi.org/10.52465/joscex.v3i1.61.
K. Jha and S. Saha, “Incorporation of multimodal multiobjective optimization in designing a filter based feature selection technique,” Appl. Soft Comput., vol. 98, p. 106823, 2021, https://doi.org/10.1016/j.asoc.2020.106823.
C. Jie, L. Jiawei, W. Shulin, and Y. Sheng, “Feature selection in machine learning: A new perspective,” Neurocomputing, vol. 300, pp. 70–79, 2018, https://doi.org/10.1016/j.neucom.2017.11.077.
R. Sheikhpour, M. A. Sarram, S. Gharaghani, and M. A. Z. Chahooki, “A survey on semi-supervised feature selection methods,” Pattern Recognit., vol. 64, pp. 141–158, 2017, https://doi.org/10.1016/j.patcog.2016.11.003.
N. Gopika and A. M. Kowshalaya. M. E, “Correlation based feature selection algorithm for machine learning,” in 2018 3rd international conference on communication and electronics systems (ICCES), pp. 692–695, 2018, https://doi.org/10.1109/CESYS.2018.8723980.
Z. Chuanlei, Z. Shanwen, Y. Jucheng, S. Yancui, and C. Jia, “Apple leaf disease identification using genetic algorithm and correlation based feature selection method,” Int. J. Agric. Biol. Eng., vol. 10, no. 2, pp. 74–83, 2017, http://www.ijabe.org/index.php/ijabe/article/view/2166.
M. Mafarja and S. Mirjalili, “Whale optimization approaches for wrapper feature selection,” Appl. Soft Comput., vol. 62, pp. 441–453, 2018, https://doi.org/10.1016/j.asoc.2017.11.006.
K. Yan, L. Ma, Y. Dai, W. Shen, Z. Ji, and D. Xie, “Cost-sensitive and sequential feature selection for chiller fault detection and diagnosis,” Int. J. Refrig., vol. 86, pp. 401–409, 2018, https://doi.org/10.1016/j.ijrefrig.2017.11.003.
A. K. Shrivas, S. K. Sahu, and H. S. Hota, “Classification of chronic kidney disease with proposed union based feature selection technique,” in Proceedings of 3rd International Conference on Internet of Things and Connected Technologies (ICIoTCT), pp. 26–27, 2018, https://doi.org/10.2139/ssrn.3168581.
I. M. Nasir et al., “Pearson correlation-based feature selection for document classification using balanced training,” Sensors, vol. 20, no. 23, p. 6793, 2020, https://doi.org/10.3390/s20236793.
F. Nojavan, S. S. Qian, and C. A. Stow, “Comparative analysis of discretization methods in Bayesian networks,” Environ. Model. Softw., vol. 87, pp. 64–71, 2017, https://doi.org/10.1016/j.envsoft.2016.10.007.
S. S. Pal and S. Kar, “Time series forecasting for stock market prediction through data discretization by fuzzistics and rule generation by rough set theory,” Math. Comput. Simul., vol. 162, pp. 18–30, 2019, https://doi.org/10.1016/j.matcom.2019.01.001.
N. Thein, K. Hamamoto, H. A. Nugroho, and T. B. Adji, “A comparison of three preprocessing techniques for kidney stone segmentation in CT scan images,” in 2018 11th Biomedical Engineering International Conference (BMEiCON), pp. 1–5, 2018, https://doi.org/10.1109/BMEiCON.2018.8609996.
P. Romagnani et al., “Chronic kidney disease,” Nat. Rev. Dis. Prim., vol. 3, no. 1, pp. 1–24, 2017, https://doi.org/10.1038/nrdp.2017.88.
Centers for Disease Control and Prevention, Chronic kidney disease in the United States, 2019. Atlanta, GA: US Department of Health and Human Services, Centers for Disease Control and Prevention, 2019, https://fluoridealert.org/studytracker/38332/.
T. K. Chen, D. H. Knicely, and M. E. Grams, “Chronic kidney disease diagnosis and management: a review,” Jama, vol. 322, no. 13, pp. 1294–1304, 2019, https://doi.org/10.1001/jama.2019.14745.
A. C. Webster, E. V Nagler, R. L. Morton, and P. Masson, “Chronic kidney disease,” Lancet, vol. 389, no. 10075, pp. 1238–1252, 2017, https://doi.org/10.1016/S0140-6736(16)32064-5.
W. Zheng et al., “Improving crop yields, nitrogen use efficiencies, and profits by using mixtures of coated controlled-released and uncoated urea in a wheat-maize system,” F. Crop. Res., vol. 205, pp. 106–115, 2017, https://doi.org/10.1016/j.fcr.2017.02.009.
J. L. Segar et al., “Fluid management, electrolytes imbalance and renal management in neonates with neonatal encephalopathy treated with hypothermia,” in Seminars in Fetal and Neonatal Medicine, vol. 26, no. 4, p. 101261, 2021, https://doi.org/10.1016/j.siny.2021.101261.
S. Javaid, H. Awais, M. Usman, and U. Mukhtar, “Biochemical Changes in Chronic Kidney Disease (CKD) Patients and its Association with Hypertension and Diabetes Mellitus,” Asian J. Allied Heal. Sci., vol. 6, no. 2, 2021, https://jucmd.pk/journals/AJAHS/article/view/1415.
K. L. Watts, P. Ghosh, S. Stein, and R. Ghavamian, “Value of nephrometry score constituents on perioperative outcomes and split renal function in patients undergoing minimally invasive partial nephrectomy,” Urology, vol. 99, pp. 112–117, 2017, https://doi.org/10.1016/j.urology.2016.01.046.
M. Liu et al., “Personal exposure to fine particulate matter and renal function in children: a panel study,” Environ. Pollut., vol. 266, p. 115129, 2020, https://doi.org/10.1016/j.envpol.2020.115129.
J. P. Kooman et al., “Inflammation and premature aging in advanced chronic kidney disease,” Am. J. Physiol. Physiol., vol. 313, no. 4, pp. F938–F950, 2017, https://doi.org/10.1152/ajprenal.00256.2017.
W. F. Clark et al., “Effect of coaching to increase water intake on kidney function decline in adults with chronic kidney disease: the CKD WIT randomized clinical trial,” Jama, vol. 319, no. 18, pp. 1870–1879, 2018, https://doi.org/10.1001/jama.2018.4930.
F. Ridzuan and W. M. N. W. Zainon, “A review on data cleansing methods for big data,” Procedia Comput. Sci., vol. 161, pp. 731–738, 2019, https://doi.org/10.1016/j.procs.2019.11.177.
C. B. Rjeily, G. Badr, A. Hajjarm El Hassani, and E. Andres, “Medical data mining for heart diseases and the future of sequential mining in medical field,” in Machine Learning Paradigms, pp. 71–99, 2019, https://doi.org/10.1007/978-3-319-94030-4_4.
I. S. Thaseen, J. Saira Banu, K. Lavanya, M. Rukunuddin Ghalib, and K. Abhishek, “An integrated intrusion detection system using correlation‐based attribute selection and artificial neural network,” Trans. Emerg. Telecommun. Technol., vol. 32, no. 2, 2021, https://doi.org/10.1002/ett.4014.
D. Chutia, D. K. Bhattacharyya, J. Sarma, and P. N. L. Raju, “An effective ensemble classification framework using random forests and a correlation based feature selection technique,” Trans. GIS, vol. 21, no. 6, pp. 1165–1178, 2017, https://doi.org/10.1111/tgis.12268.
F. Hamedan, A. Orooji, H. Sanadgol, and A. Sheikhtaheri, “Clinical decision support system to predict chronic kidney disease: A fuzzy expert system approach,” Int. J. Med. Inform., vol. 138, p. 104134, 2020, https://doi.org/10.1016/j.ijmedinf.2020.104134.
N. Cahyani and M. A. Muslim, “Increasing Accuracy of C4. 5 Algorithm by applying discretization and correlation-based feature selection for chronic kidney disease diagnosis,” Journal of Telecommunication, Electronic and Computer Engineering (JTEC), vol. 12, no. 1, pp. 25-32, 2020, https://jtec.utem.edu.my/jtec/article/view/4922.
N. A. Almansour et al., “Neural network and support vector machine for the prediction of chronic kidney disease: A comparative study,” Comput. Biol. Med., vol. 109, pp. 101–111, 2019, https://doi.org/10.1016/j.compbiomed.2019.04.017.
Published
How to Cite
Issue
Section
License
Copyright (c) 2023 Dwika Ananda Agustina Pertiwi, Pipit Riski Setyorini, Much Aziz Muslim, Endang Sugiharti

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
Authors who publish with this journal agree to the following terms:
- Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this journal.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial publication in this journal.
- Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See The Effect of Open Access).
This journal is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.


