Enhancing Early Diabetes Detection Using Tree-Based Machine Learning Algorithms with SMOTEENN Balancing
DOI:
https://doi.org/10.12928/mf.v8i1.14495Keywords:
diabetes, detection, Machine Learning, Classification, data balancingAbstract
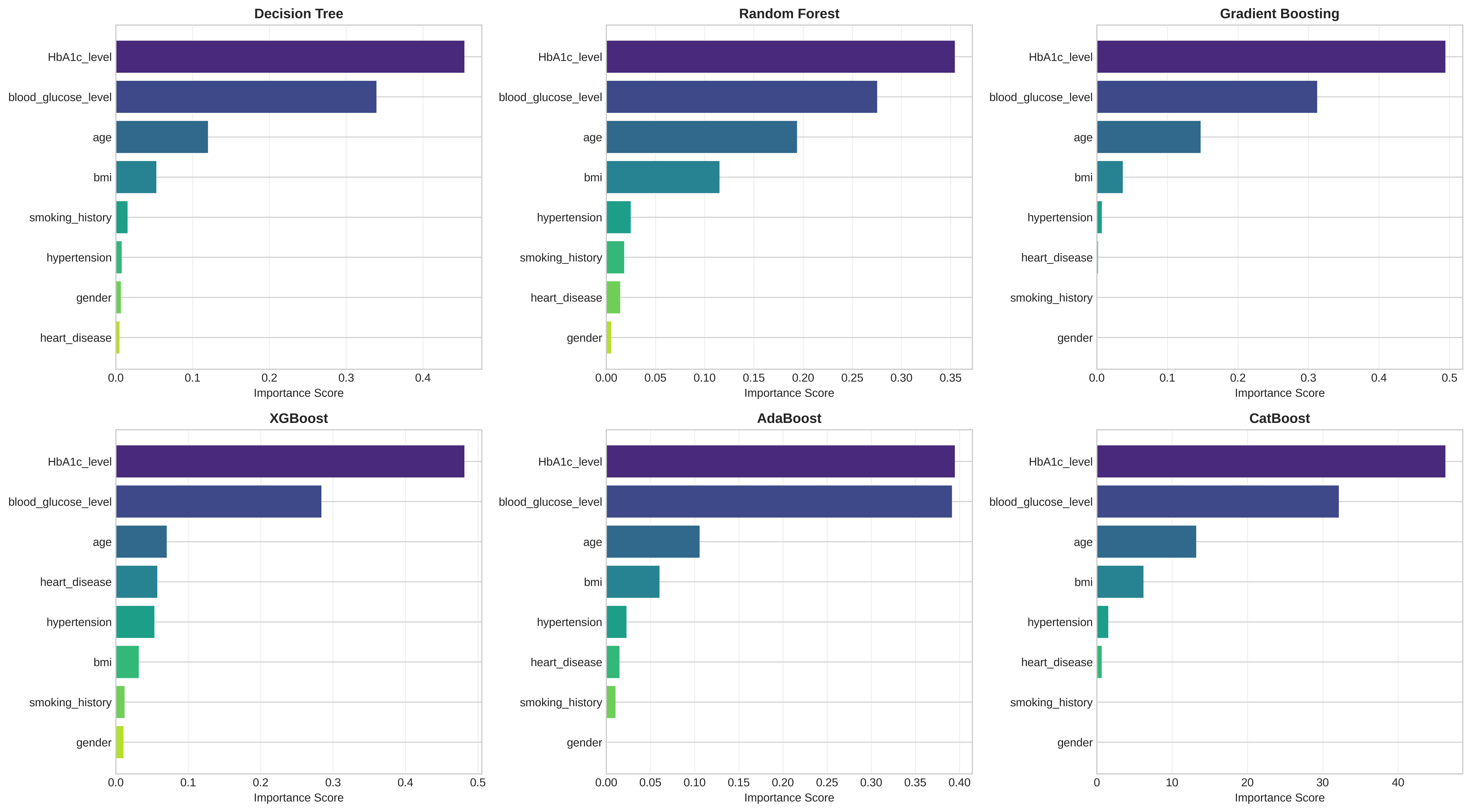
Diabetes continues to be a critical global health issue, demanding accurate predictive systems to enable preventive interventions. Traditional diagnostic tests lack efficiency for large-scale early screening, which has led to growing interest in artificial intelligence solutions. This research proposed an effective methodology for diabetes classification based on tree-based algorithms enhanced with SMOTEENN balancing. The study employed the Kaggle Diabetes Prediction Dataset with 100,000 instances and eight medical and demographic features. Preprocessing steps included handling missing and duplicate values, encoding categorical variables, and scaling numerical attributes with Min-Max normalization. To address severe class imbalance, SMOTEENN was adopted, producing a cleaner and more balanced dataset. Model evaluation was performed using Stratified 5-Fold cross-validation on six classifiers: Decision Tree, Random Forest, Gradient Boosting, AdaBoost, XGBoost, and CatBoost. Experimental results indicated significant gains after balancing, with ensemble methods outperforming single-tree baselines. Random Forest delivered the best overall performance (98.93% accuracy, 98.96% F1-score, 99.16% recall, 99.94% AUC), followed by CatBoost and XGBoost with comparable results above 99% AUC. While Decision Tree benefited most from SMOTEENN in relative terms, it remained less competitive. Analysis of the importance of the analysis revealed HbA1c level and blood glucose level as dominant predictors, validating clinically meaningful learning. These findings suggest that integrating hybrid resampling with ensemble tree classifiers provides reliable and general predictions for diabetes risk. The approach holds promise for deployment in healthcare decision support systems.
References
L. A. Al Hak, “Diabetes Prediction Using Binary Grey Wolf Optimization and Decision Tree,” Int. J. Comput., vol. 21, no. 4, pp. 489–494, 2022, doi: 10.47839/ijc.21.4.2785.
S. Sivaranjani, S. Ananya, J. Aravinth, and R. Karthika, “Diabetes Prediction using Machine Learning Algorithms with Feature Selection and Dimensionality Reduction,” in 2021 7th International Conference on Advanced Computing and Communication Systems (ICACCS), IEEE, Mar. 2021, pp. 141–146. doi: 10.1109/ICACCS51430.2021.9441935.
M. M. Bukhari, B. F. Alkhamees, S. Hussain, A. Gumaei, A. Assiri, and S. S. Ullah, “An Improved Artificial Neural Network Model for Effective Diabetes Prediction,” Complexity, vol. 2021, no. 1, Jan. 2021, doi: 10.1155/2021/5525271.
S. M. Ganie and M. B. Malik, “An ensemble Machine Learning approach for predicting Type-II diabetes mellitus based on lifestyle indicators,” Healthc. Anal., vol. 2, no. January, p. 100092, 2022, doi: 10.1016/j.health.2022.100092.
M. Revathi, A. B. Godbin, S. N. Bushra, and S. Anslam Sibi, “Application of ANN, SVM and KNN in the Prediction of Diabetes Mellitus,” in 2022 International Conference on Electronic Systems and Intelligent Computing (ICESIC), IEEE, Apr. 2022, pp. 179–184. doi: 10.1109/ICESIC53714.2022.9783577.
N. G. Ramadhan, “Comparative Analysis of ADASYN-SVM and SMOTE-SVM Methods on the Detection of Type 2 Diabetes Mellitus,” Sci. J. Informatics, vol. 8, no. 2, pp. 276–282, 2021, doi: 10.15294/sji.v8i2.32484.
A. Fauzi and A. H. Yunial, “Optimasi Algoritma Klasifikasi Naive Bayes, Decision Tree, K – Nearest Neighbor, dan Random Forest menggunakan Algoritma Particle Swarm Optimization pada Diabetes Dataset,” J. Edukasi dan Penelit. Inform., vol. 8, no. 3, p. 470, Dec. 2022, doi: 10.26418/jp.v8i3.56656.
P. Sugiartawan, N. W. Wardani, A. A. S. Pradhana, K. S. Batubulan, and I. N. D. Kotama, “Support Vector Machine for Accurate Classification of Diabetes Risk Levels,” IJCCS (Indonesian J. Comput. Cybern. Syst., vol. 19, no. 3, pp. 317–326, Jul. 2025, doi: 10.22146/ijccs.107740.
A. Dutta et al., “Early Prediction of Diabetes Using an Ensemble of Machine Learning Models,” Int. J. Environ. Res. Public Health, vol. 19, no. 19, p. 12378, Sep. 2022, doi: 10.3390/ijerph191912378.
M. T. García-Ordás, C. Benavides, J. A. Benítez-Andrades, H. Alaiz-Moretón, and I. García-Rodríguez, “Diabetes detection using deep learning techniques with oversampling and feature augmentation,” Comput. Methods Programs Biomed., vol. 202, p. 105968, Apr. 2021, doi: 10.1016/j.cmpb.2021.105968.
B. Aruna Devi and N. Karthik, “The Effect of Anomaly Detection and Data Balancing in Prediction of Diabetes,” Int. J. Comput. Inf. Syst. Ind. Manag. Appl., vol. 16, no. 2, pp. 184–196, 2024.
Kartina Diah Kusuma Wardani and Memen Akbar, “Diabetes Risk Prediction using Feature Importance Extreme Gradient Boosting (XGBoost),” J. RESTI (Rekayasa Sist. dan Teknol. Informasi), vol. 7, no. 4, pp. 824–831, Aug. 2023, doi: 10.29207/resti.v7i4.4651.
A. Wibowo, A. F. N. Masruriyah, and S. Rahmawati, “Refining Diabetes Diagnosis Models: The Impact of SMOTE on SVM, Logistic Regression, and Naïve Bayes,” J. Electron. Electromed. Eng. Med. Informatics, vol. 7, no. 1, pp. 197–207, Jan. 2025, doi: 10.35882/jeeemi.v7i1.596.
V. Sinap, “The Impact of Balancing Techniques and Feature Selection on Machine Learning Models for Diabetes Detection,” Fırat Üniversitesi Mühendislik Bilim. Derg., vol. 37, no. 1, pp. 303–320, Mar. 2025, doi: 10.35234/fumbd.1556260.
S. Feng, J. Keung, X. Yu, Y. Xiao, and M. Zhang, “Investigation on the stability of SMOTE-based oversampling techniques in software defect prediction,” Inf. Softw. Technol., vol. 139, p. 106662, 2021, doi: 10.1016/j.infsof.2021.106662.
A. S. Barkah, S. R. Selamat, Z. Z. Abidin, and R. Wahyudi, “Impact of Data Balancing and Feature Selection on Machine Learning-based Network Intrusion Detection,” Int. J. Informatics Vis., vol. 7, no. 1, pp. 241–248, 2023, doi: 10.30630/joiv.7.1.1041.
C. C. Olisah, L. Smith, and M. Smith, “Diabetes mellitus prediction and diagnosis from a data preprocessing and machine learning perspective,” Comput. Methods Programs Biomed., vol. 220, p. 106773, 2022, doi: 10.1016/j.cmpb.2022.106773.
S. Alam and N. Yao, “The impact of preprocessing steps on the accuracy of machine learning algorithms in sentiment analysis,” Comput. Math. Organ. Theory, vol. 25, pp. 319–335, 2019, doi: 10.1007/s10588-018-9266-8.
I. Riadi, A. Yudhana, and M. R. Djou, “Optimization of Population Document Services in Villages using Naive Bayes and k-NN Method,” Int. J. Comput. Digit. Syst., vol. 1, no. 1, pp. 127–138, 2024, doi: 10.12785/ijcds/150111.
N. Alnor, A. Khleel, and K. Nehéz, “Improving the accuracy of recurrent neural networks models in predicting software bug based on undersampling methods,” Indones. J. Electr. Eng. Comput. Sci., vol. 32, no. 1, pp. 478–493, 2023, doi: 10.11591/ijeecs.v32.i1.pp478-493.
H. F. El-Sofany, “Predicting Heart Diseases Using Machine Learning and Different Data Classification Techniques,” IEEE Access, vol. 12, no. July, pp. 106146–106160, 2024, doi: 10.1109/ACCESS.2024.3437181.
A. Luque, A. Carrasco, A. Martín, and A. de las Heras, “The impact of class imbalance in classification performance metrics based on the binary confusion matrix,” Pattern Recognit., vol. 91, pp. 216–231, Jul. 2019, doi: 10.1016/j.patcog.2019.02.023.
V. Kumar, G. S. Lalotra, and R. K. Kumar, “Improving performance of classifiers for diagnosis of critical diseases to prevent COVID risk,” Comput. Electr. Eng., vol. 102, p. 108236, Sep. 2022, doi: 10.1016/j.compeleceng.2022.108236.
C. Yu, Y. Jin, Q. Xing, Y. Zhang, S. Guo, and S. Meng, “Advanced User Credit Risk Prediction Model Using LightGBM, XGBoost and Tabnet with SMOTEENN,” 2024 IEEE 6th Int. Conf. Power, Intell. Comput. Syst. ICPICS 2024, pp. 876–883, 2024, doi: 10.1109/ICPICS62053.2024.10796247.
E. Mbunge et al., “Implementation of ensemble machine learning classifiers to predict diarrhoea with SMOTEENN, SMOTE, and SMOTETomek class imbalance approaches,” 2023 Conf. Inf. Commun. Technol. Soc. ICTAS 2023 - Proc., 2023, doi: 10.1109/ICTAS56421.2023.10082744.
H. A. Abdelhafez, “Machine Learning Techniques for Diabetes Prediction: A Comparative Analysis,” J. Appl. Data Sci., vol. 5, no. 2, pp. 792–807, May 2024, doi: 10.47738/jads.v5i2.219.
S. Saxena, D. Mohapatra, S. Padhee, and G. K. Sahoo, “Machine learning algorithms for diabetes detection: a comparative evaluation of performance of algorithms,” Evol. Intell., vol. 16, no. 2, pp. 587–603, Apr. 2023, doi: 10.1007/s12065-021-00685-9.
K. Abnoosian, R. Farnoosh, and M. H. Behzadi, “Prediction of diabetes disease using an ensemble of machine learning multi-classifier models,” BMC Bioinformatics, vol. 24, no. 1, pp. 1–24, 2023, doi: 10.1186/s12859-023-05465-z.
K. D. K. Wardhani and M. Akbar, “Diabetes Risk Prediction Using Extreme Gradient Boosting (XGBoost),” J. Online Inform., vol. 7, no. 2, pp. 244–250, Dec. 2022, doi: 10.15575/join.v7i2.970.
V. Chang, J. Bailey, Q. A. Xu, and Z. Sun, “Pima Indians diabetes mellitus classification based on machine learning (ML) algorithms,” Neural Comput. Appl., vol. 35, no. 22, pp. 16157–16173, Aug. 2023, doi: 10.1007/s00521-022-07049-z.
A. Vanacore, M. S. Pellegrino, and A. Ciardiello, “Fair evaluation of classifier predictive performance based on binary confusion matrix,” Comput. Stat., Nov. 2022, doi: https://doi.org/10.1007/s00180-022-01301-9.
D. K. Sharma, M. Chatterjee, G. Kaur, and S. Vavilala, “Deep learning applications for disease diagnosis,” in Deep Learning for Medical Applications with Unique Data, Elsevier, 2022, pp. 31–51. doi: 10.1016/B978-0-12-824145-5.00005-8.
H. Abbad Ur Rehman, C. Y. Lin, and Z. Mushtaq, “Effective K-Nearest Neighbor Algorithms Performance Analysis of Thyroid Disease,” J. Chinese Inst. Eng. Trans. Chinese Inst. Eng. A, vol. 44, no. 1, pp. 77–87, 2021, doi: 10.1080/02533839.2020.1831967.
P. Singh, N. Singh, K. K. Singh, and A. Singh, “Diagnosing of disease using machine learning,” Mach. Learn. Internet Med. Things Healthc., pp. 89–111, 2021, doi: 10.1016/B978-0-12-821229-5.00003-3.
Z. Mushtaq, A. Yaqub, S. Sani, and A. Khalid, “Effective K-nearest neighbor classifications for Wisconsin breast cancer data sets,” J. Chinese Inst. Eng. Trans. Chinese Inst. Eng. A, vol. 43, no. 1, pp. 80–92, 2020, doi: 10.1080/02533839.2019.1676658.
Downloads
Published
Issue
Section
License
Copyright (c) 2025 Syahrani Lonang, Ahmad Fatoni Dwi Putra, Asno Azzawagama Firdaus, Fahmi Syuhada, Yuan Sa'adati

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
Start from 2019 issues, authors who publish with JURNAL MOBILE AND FORENSICS agree to the following terms:
- Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution License (CC BY-SA 4.0) that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this journal.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial publication in this journal.
- Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work.

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.












 Mobile and Forensics (MF)
Mobile and Forensics (MF)