Identification of Plasmodium Vivax in Blood Smear Images Using Otsu Thresholding Algorithm
DOI:
https://doi.org/10.12928/mf.v6i1.11261Keywords:
CNN, Malaria, Plasmodium Vivax, SVM, Otsu ThresholdingAbstract
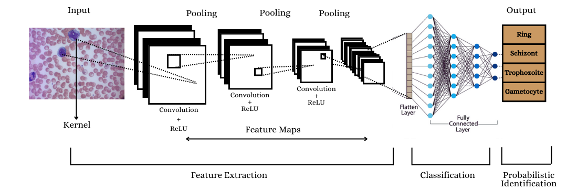
In this research, we explore the efficacy of Convolutional Neural Networks (CNN) and Support Vector Machines (SVM) in identifying Plasmodium vivax from blood smear images. We utilized a dataset comprising images of Plasmodium vivax and non-infected cells, applying CNN for deep feature extraction and SVM with otsu’s thresholding for segmentation. The dataset was preprocessed and augmented to enhance model performance. The CNN architecture, consisting of multiple convolutional and dense layers, achieved an accuracy of 98.46% on the validation set. For comparison, features extracted using Otsu’s Thresholding were fed into an SVM classifier, yielding an accuracy of 82%. Confusion matrix was generated to evaluate the classification performance of both models. The CNN model demonstrated superior accuracy and robustness in classification tasks compared to the SVM model. This research shows how deep learning frameworks can be used to analyse medical images and how important it is to have methods for extracting and choosing features to make machine learning models work better.
References
I. M. D. Maysanjaya, “Comparative study of classification method on diagnosis of plasmodium phase,” J. Phys. Conf. Ser., vol. 1516, no. 1, 2020, doi: 10.1088/1742-6596/1516/1/012021.
M. Fu, K. Wu, Y. Li, L. Luo, W. Huang, and Q. Zhang, “An intelligent detection method for plasmodium based on self-supervised learning and attention mechanism,” Front. Med., vol. 10, no. June, pp. 1–8, 2023, doi: 10.3389/fmed.2023.1117192.
N. Huda, A. Y. Dewi, and A. Mahiruna, “Plasmodium falciparum Identification Using Otsu Thresholding Segmentation Method Based on Microscopic Blood Image,” vol. 10, no. 4, pp. 479–488, 2023, doi: 10.15294/sji.v10i4.47924.
D. Crossed D Signumic, D. Keco, and Z. Mašetic, “Automatization of Microscopy Malaria Diagnosis Using Computer Vision and Random Forest Method,” IFAC-PapersOnLine, vol. 55, no. 4, pp. 80–84, 2022, doi: 10.1016/j.ifacol.2022.06.013.
D. O. Oyewola, E. G. Dada, S. Misra, and R. Damaševičius, “A Novel Data Augmentation Convolutional Neural Network for Detecting Malaria Parasite in Blood Smear Images,” Appl. Artif. Intell., vol. 36, no. 1, 2022, doi: 10.1080/08839514.2022.2033473.
A. de Souza Oliveira, M. Guimarães Fernandes Costa, M. das Graças Vale Barbosa, and C. Ferreira Fernandes Costa Filho, “A new approach for malaria diagnosis in thick blood smear images,” Biomed. Signal Process. Control, vol. 78, no. April, 2022, doi: 10.1016/j.bspc.2022.103931.
R. Saikia and S. S. Devi, “White Blood Cell Classification based on Gray Level Co-occurrence Matrix with Zero Phase Component Analysis Approach,” Procedia Comput. Sci., vol. 218, pp. 1977–1984, 2023, doi: 10.1016/j.procs.2023.01.174.
A. Loddo, C. Fadda, and C. Di Ruberto, “An Empirical Evaluation of Convolutional Networks for Malaria Diagnosis,” J. Imaging, vol. 8, no. 3, 2022, doi: 10.3390/jimaging8030066.
M. Abdel-Basset, R. Mohamed, M. Abouhawwash, S. S. Askar, and A. A. Tantawy, “An Efficient Multilevel Threshold Segmentation Method for Breast Cancer Imaging Based on Metaheuristics Algorithms: Analysis and Validations,” Int. J. Comput. Intell. Syst., vol. 16, no. 1, 2023, doi: 10.1007/s44196-023-00282-x.
P. A. Pitoy and I. P. G. H. Suputra, “Dermoscopy Image Segmentation in Melanoma Skin Cancer using Otsu Thresholding Method,” JELIKU (Jurnal Elektron. Ilmu Komput. Udayana), vol. 9, no. 3, p. 397, 2021, doi: 10.24843/jlk.2021.v09.i03.p11.
M. Wattana and T. Boonsri, “Improvement of complete malaria cell image segmentation,” 2017 12th Int. Conf. Digit. Inf. Manag. ICDIM 2017, vol. 2018-Janua, no. Icdim, pp. 319–323, 2018, doi: 10.1109/ICDIM.2017.8244655.
H. A. Nugroho, I. M. Dendi Maysanjaya, N. A. Setiawan, E. E. H. Murhandarwati, and W. K. Z. Oktoeberza, “Feature analysis for stage identification of Plasmodium vivax based on digital microscopic image,” Indones. J. Electr. Eng. Comput. Sci., vol. 13, no. 2, pp. 721–728, 2019, doi: 10.11591/ijeecs.v13.i2.pp721-728.
D. Nauriyal and D. Kumar, “Study of Severe Malaria Caused by Plasmodium Vivax in Comparison to Plasmodium Falciparum and Mixed Malarial Infections in Children,” Biomed. Pharmacol. J., vol. 15, no. 3, pp. 1597–1604, 2022, doi: 10.13005/bpj/2498.
L. Muflikhah, N. Widodo, N. Yudistira, and A. Ridok, “Drug Resistant Prediction Based on Plasmodium Falciparum DNA-Barcoding using Bidirectional Long Short Term Memory Method,” Int. J. Adv. Comput. Sci. Appl., vol. 14, no. 7, pp. 433–440, 2023, doi: 10.14569/IJACSA.2023.0140747.
D. A. L. Kafaf, N. N. Thamir, and S. S. Al-Hadithy, “Malaria Disease Prediction Based on Convolutional Neural Networks,” J. Appl. Eng. Technol. Sci., vol. 5, no. 2, pp. 1165–1181, 2024, doi: 10.37385/jaets.v5i2.3947.
V. Magotra and M. K. Rohil, “Malaria Diagnosis Using a Lightweight Deep Convolutional Neural Network,” Int. J. Telemed. Appl., vol. 2022, 2022, doi: 10.1155/2022/4176982.
A. Dev, M. M. Fouda, L. Kerby, and Z. Md Fadlullah, “Advancing Malaria Identification from Microscopic Blood Smears Using Hybrid Deep Learning Frameworks,” IEEE Access, vol. 12, no. May, pp. 71705–71715, 2024, doi: 10.1109/ACCESS.2024.3402442.
S. Pal, R. P. Singh, and A. Kumar, “Analysis of Hybrid Feature Optimization Techniques Based on the Classification Accuracy of Brain Tumor Regions Using Machine Learning and Further Evaluation Based on the Institute Test Data,” J. Med. Phys., vol. 49, no. 1, pp. 22–32, 2024, doi: 10.4103/jmp.jmp_77_23.
A. Vijayalakshmi and R. Kanna B, “Deep learning approach to detect malaria from microscopic images,” Multimed. Tools Appl., vol. 79, no. 21, pp. 15297–15317, 2020, doi: 10.1007/s11042-019-7162-y.
H. K. Ragb, I. T. Dover, and R. Ali, “Deep convolutional neural network ensemble for improved malaria parasite detection,” Proc. - Appl. Imag. Pattern Recognit. Work., vol. 2020-Octob, 2020, doi: 10.1109/AIPR50011.2020.9425273.
C. B. Jones and C. Murugamani, “Malaria Parasite Detection on Microscopic Blood Smear Images with Integrated Deep Learning Algorithms,” Int. Arab J. Inf. Technol., vol. 20, no. 2, pp. 170–179, 2023, doi: 10.34028/iajit/20/2/3.
M. Yebasse, K. J. Cheoi, and J. Ko, “Malaria Disease Cell Classification With Highlighting Small Infected Regions,” IEEE Access, vol. 11, no. January, pp. 15945–15953, 2023, doi: 10.1109/ACCESS.2023.3245025.
D. Setyawan, R. Wardoyo, M. E. Wibowo, and E. E. H. Murhandarwati, “Classification of plasmodium falciparum based on textural and morphological features,” Int. J. Electr. Comput. Eng., vol. 12, no. 5, pp. 5036–5048, 2022, doi: 10.11591/ijece.v12i5.pp5036-5048.
J. Liao, Y. Wang, D. Zhu, Y. Zou, S. Zhang, and H. Zhou, “Automatic Segmentation of Crop/Background Based on Luminance Partition Correction and Adaptive Threshold,” IEEE Access, vol. 8, pp. 202611–202622, 2020, doi: 10.1109/ACCESS.2020.3036278.
Singh;, M. Mehra, A. Kumar, M. . Niranjannaik, D. Priya, and K. Gaurav, “Leveraging hybrid machine learning and data fusion for accurate mapping of malaria cases using meteorological variables in western India,” Intell. Syst. with Appl., vol. 17, no. February 2022, p. 200164, 2023, doi: 10.1016/j.iswa.2022.200164.
G. Zhang et al., “Adaptive threshold model in google earth engine: A case study of ulva prolifera extraction in the south yellow sea, China,” Remote Sens., vol. 13, no. 16, 2021, doi: 10.3390/rs13163240.
M. H. Malik, H. Ghous, T. Rashid, B. Maryum, Z. Hao, and Q. Umer, “Feature extraction-based liver tumor classification using Machine Learning and Deep Learning methods of computed tomography images,” Cogent Eng., vol. 11, no. 1, p., 2024, doi: 10.1080/23311916.2024.2338994.
H. A. Nugroho, M. S. Wibawa, N. A. Setiawan, E. E. H. Murhandarwati, and R. L. B. Buana, “Identification of Plasmodium falciparum and Plasmodium vivax on digital image of thin blood films,” Indones. J. Electr. Eng. Comput. Sci., vol. 13, no. 3, pp. 933–944, 2019, doi: 10.11591/ijeecs.v13.i3.pp933-944.
S. N. A. M. Kanafiah, M. Y. Mashor, Z. Mohamed, Y. C. Way, S. A. A. Shukor, and Y. Jusman, “An Intelligent Classification System for Trophozoite Stages in Malaria Species,” Intell. Autom. Soft Comput., vol. 34, no. 1, pp. 687–697, 2022, doi: 10.32604/iasc.2022.024361.
Y. Liu, F. C. Lin, J. T. Lin, and Q. Li, “Dynamic Classification of Plasmodium vivax Malaria Recurrence: An Application of Classifying Unknown Cause of Failure in Competing Risks,” J. Data Sci., vol. 20, no. 1, pp. 51–78, 2022, doi: 10.6339/21-JDS1026.
S. Suganyadevi, V. Seethalakshmi, and K. Balasamy, “A review on deep learning in medical image analysis,” Int. J. Multimed. Inf. Retr., vol. 11, no. 1, pp. 19–38, 2022, doi: 10.1007/s13735-021-00218-1.
K. E. D. Penas, P. T. Rivera, and P. C. Naval, “Malaria Parasite Detection and Species Identification on Thin Blood Smears Using a Convolutional Neural Network,” Proc. - 2017 IEEE 2nd Int. Conf. Connect. Heal. Appl. Syst. Eng. Technol. CHASE 2017, pp. 1–6, 2017, doi: 10.1109/CHASE.2017.51.
A. Bin Abdul Qayyum, T. Islam, and M. A. Haque, “Malaria Diagnosis with Dilated Convolutional Neural Network Based Image Analysis,” BECITHCON 2019 - 2019 IEEE Int. Conf. Biomed. Eng. Comput. Inf. Technol. Heal., pp. 68–72, 2019, doi: 10.1109/BECITHCON48839.2019.9063179.
Downloads
Published
Issue
Section
License
Copyright (c) 2024 Nurul Huda, Aulia, Citra

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
Start from 2019 issues, authors who publish with JURNAL MOBILE AND FORENSICS agree to the following terms:
- Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution License (CC BY-SA 4.0) that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this journal.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial publication in this journal.
- Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work.

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.












 Mobile and Forensics (MF)
Mobile and Forensics (MF)