ISSN: 2685-9572 Buletin Ilmiah Sarjana Teknik Elektro
Vol. 7, No. 3, September 2025, pp. 541-553
Transfer Learning Models for Precision Medicine: A Review of Current Applications
Yuri Pamungkas 1, Myo Min Aung 2, Gao Yulan 3, Muhammad Nur Afnan Uda 4, Uda Hashim 5
1 Department of Medical Technology, Institut Teknologi Sepuluh Nopember, Indonesia
2 Department of Mechatronics Engineering, Rajamangala University of Technology Thanyaburi, Thailand
3 Department of Mechanical Engineering, Guizhou University of Engineering Science, China
4, 5 Department of Electrical Electronic Engineering, Universiti Malaysia Sabah, Malaysia
ARTICLE INFORMATION |
| ABSTRACT |
Article History: Received 17 July 2025 Revised 19 August 2025 Accepted 25 September 2025 |
| 
In recent years, Transfer Learning (TL) models have demonstrated significant promise in advancing precision medicine by enabling the application of machine learning techniques to medical data with limited labeled information. TL overcomes the challenge of acquiring large, labeled datasets, which is often a limitation in medical fields. By leveraging knowledge from pre-trained models, TL offers a solution to improve diagnostic accuracy and decision-making processes in various healthcare domains, including medical imaging, disease classification, and genomics. The research contribution of this review is to systematically examine the current applications of TL models in precision medicine, providing insights into how these models have been successfully implemented to improve patient outcomes across different medical specialties. In this review, studies sourced from the Scopus database, all published in 2024 and selected for their "open access" availability, were analyzed. The research methods involved using TL techniques like fine-tuning, feature-based learning, and model-based transfer learning on diverse datasets. The results of the studies demonstrated that TL models significantly enhanced the accuracy of medical diagnoses, particularly in areas such as brain tumor detection, diabetic retinopathy, and COVID-19 detection. Furthermore, these models facilitated the classification of rare diseases, offering valuable contributions to personalized medicine. In conclusion, Transfer Learning has the potential to revolutionize precision medicine by providing cost-effective and scalable solutions for improving diagnostic capabilities and treatment personalization. The continued development and integration of TL models in clinical practice promise to further enhance the quality of patient care. |
Keywords: Transfer Learning; Precision Medicine; Medical Imaging; Deep Learning; Personalized Healthcare |
Corresponding Author: Yuri Pamungkas, Department of Medical Technology, Institut Teknologi Sepuluh Nopember, Indonesia. Email: yuri@its.ac.id |
This work is open access under a Creative Commons Attribution-Share Alike 4.0 
|
Document Citation: Y. Pamungkas, M. M. Aung, G. Yulan, M. N. A. Uda, and U. Hashim, “Transfer Learning Models for Precision Medicine: A Review of Current Applications,” Buletin Ilmiah Sarjana Teknik Elektro, vol. 3, no. 1, pp. 541-553, 2025, DOI: 10.12928/biste.v7i3.14286. |
INTRODUCTION
Precision medicine seeks to deliver tailored healthcare grounded in personal attributes such as genetics, lifestyle, and environment [1]. However, one of the significant challenges in implementing precision medicine is the restricted access to high-quality, extensive data collections [2]. While genomic data, clinical records, and medical imaging hold valuable insights for personalized treatment, the scarcity of comprehensively labeled information hinders the development of robust predictive models [3]. Traditional machine learning (ML) methods often struggle to generalize across patient populations and disease types due to data limitations [4]. Furthermore, protection of sensitive information along with regulatory issues further complicate obtaining and distributing healthcare information across institutions.
To address these difficulties, Transfer Learning (TL) has arisen as a viable approach. TL leverages knowledge learned from a large, well-curated dataset to improve the performance of models trained on smaller, more specialized datasets [5]. In the context of precision medicine, TL allows for the reuse of pretrained models across different medical domains, including genomics, health-related imaging, and evaluation of clinical datasets [6]. This strategy is especially beneficial for addressing the scarcity of data by enabling knowledge transfer from related tasks or datasets. By applying TL, models can improve diagnostic accuracy, reduce training time, and facilitate the integration of multi-modal data for better clinical decision processes in healthcare [7].
Recent progress in TL methodologies has greatly enhanced the forefront of medical AI. Researchers have effectively implemented TL across multiple healthcare fields, such as genomics, in which pretrained models on extensive genomic datasets are adapted to target particular categories of diseases [8]. In medical imaging, TL has been used to augment small datasets, enabling improved lesion detection and image segmentation [9]. Additionally, TL has shown promise in integrating multi-omics data, where models initially developed on a single omics dataset (for example in genomics) are repurposed for another domain (for instance proteomics or metabolomics) [10]. These breakthroughs are setting the stage for more accurate and scalable solutions in precision medicine.
The originality of this study resides in its comprehensive exploration of TL applications across multiple domains of precision medicine, synthesizing recent findings to highlight the most promising strategies for model development. The contribution of this review is twofold: first, it consolidates the current state of TL applications in precision medicine; and second, it identifies the key challenges that remain, such as negative transfer and data heterogeneity. Furthermore, it provides insight into future directions that could enable further advancements, such as federated learning and the use of self-supervised models for pretraining on unlabeled medical data. By summarizing the cutting-edge applications of TL in precision medicine and pointing out the challenges and future research areas, this review aims to guide the development of more efficient, scalable, and adaptable machine learning models for personalized healthcare.
- CONCEPT OF TRANSFER LEARNING IN PRECISION MEDICINE
Transfer Learning (TL) represents an artificial intelligence method which allows models or features derived from large, general datasets to be adapted and applied to smaller, task-specific datasets [11]. This method proves especially advantageous in precision medicine, since obtaining extensive annotated datasets is frequently difficult and expensive [12]. By shifting insights from a data-rich source domain to a target domain constrained by limited information, TL enables the development of accurate and efficient models without requiring extensive data collection for the target task [13]. This is particularly crucial in medical fields where datasets can be sparse and expensive, for instance in uncommon diseases or distinct patient cohorts [14]. Additionally, TL can help overcome the challenges of data heterogeneity in medical domains, where datasets can vary significantly due to differences in population characteristics, healthcare systems, or even data collection methods [15]. By transferring knowledge across related domains, TL allows models to better generalize and adapt to the nuances of diverse patient groups, ensuring that predictive models remain robust and effective when applied to different clinical contexts [16]. This flexibility is crucial to enhance diagnostic precision and refine individualized therapeutic approaches in real-world medical settings [17].
- Target Data Types
When dealing with labeled data, Transfer Learning methods often fall under inductive transfer learning. This approach is used when the model is applied to a target domain where both input data and labeled outputs are available [18]. In precision medicine, this can be seen in applications such as disease diagnosis or patient outcome prediction where existing datasets (e.g., from clinical trials) contain both the inputs (patient data) and the corresponding outputs (disease labels or clinical outcomes). Inductive Transfer Learning enables models to generalize from these datasets to predict on unseen medical data with higher accuracy [19]. On the other hand, unannotated datasets within healthcare fields necessitate unsupervised transfer learning. This technique is employed when the target domain lacks labeled data but still contains useful input data (e.g., raw medical images or genomic data) [20]. The main objective is to derive informative attributes or abstractions out of these datasets without the necessity for explicit annotations, which can be particularly useful in genomics and medical imaging scenarios in which labeled information is frequently limited [21]. Furthermore, transductive transfer learning is implemented when a model is shifted to a related domain but where the target data may be partially labeled or consists of a small number of labeled samples, helping to fine-tune the model with minimal labeled data [22].
- Methods in Transfer Learning
In the context of Methods in Transfer Learning, various techniques can be applied to achieve better performance in precision medicine. These methods can be categorized as follows:
- Instance-based Transfer Learning focuses on relocating particular data samples from the origin domain to the destination domain [23]. This method works well in medical applications where specific case histories from one disease or treatment may be applied to similar new cases.
- Feature-based Transfer Learning aims to align and transfer feature representations across domains [24]. In precision medicine, this method is useful for tasks like medical image analysis, where the features of one set of images (e.g., from one hospital or machine) are transferred to improve the analysis of another set with different characteristics (e.g., from another hospital or imaging modality).
- Model-based Transfer Learning involves relocating the fully developed model from the origin domain to the destination domain [25]. This approach is particularly effective in scenarios like disease prediction or drug discovery, where deep learning algorithms developed using extensive datasets (e.g., gene expression data) are applied to smaller, domain-specific medical datasets.
- Fine-tuning knowledge transfer learning adjusts previously trained algorithms to the target domain’s data [26]. In precision medicine, this is widely used where an already developed model built upon an extensive dataset (e.g., general patient data) is further optimized with a narrower and more targeted dataset (e.g., a particular cancer type or a region with unique healthcare characteristics).
Every one of these methods serves a crucial function in enabling the adaptability of models to diverse medical data, facilitating the accurate prediction of disease outcomes, patient care, and therapeutic interventions, even in the face of limited labeled data [27]. Thus, Transfer Learning demonstrates strong potential in advancing precision medicine through enhancing computational effectiveness and forecast precision in healthcare applications can be seen in Figure 1 [28].
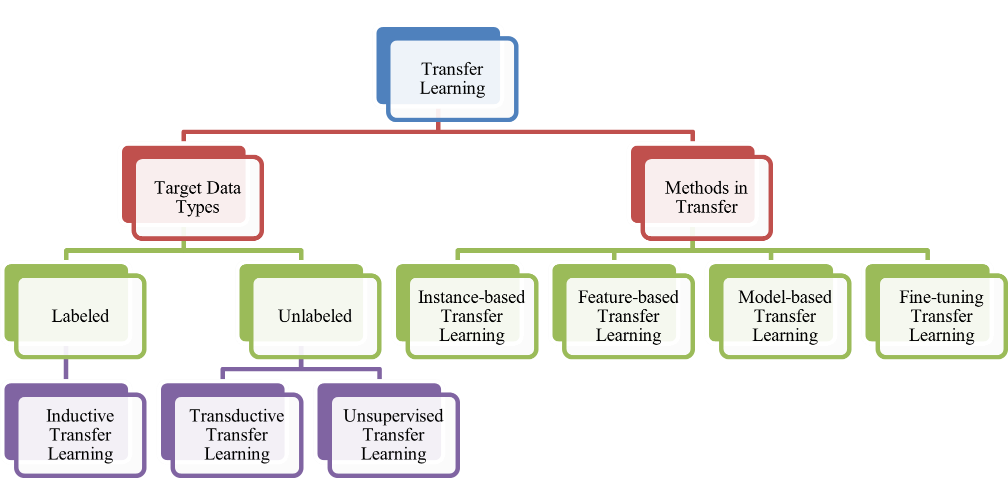
Figure 1. Types of Transfer Learning Models
- CURRENT APPLICATIONS IN PRECISION MEDICINE
The application of Transfer Learning (TL) in precision medicine has attracted considerable focus in the past few years, demonstrating its capacity to enhance medical results by providing more accurate diagnoses, personalized treatments, and enhanced decision-making processes [29][30]. This section reviews the current applications of TL models in precision medicine, drawing insights from recent research. The articles used in this review were sourced from the Scopus database, with a focus on publications released in 2024. Only "open access" articles were selected to guarantee openness and clarity of the study. To identify relevant studies, the following search query was used ("transfer learning" OR "pretrained model" OR "fine-tuning") AND ("precision medicine" OR "personalized medicine") AND ("machine learning" OR "deep learning" OR "artificial intelligence"). The search results yielded a wide range of studies that demonstrate how TL techniques are applied across various areas of precision medicine, including medical imaging, genomics, disease classification, and clinical data analysis. The following are journal articles that discuss applications of transfer learning models in precision medicine which are presented in Table 1 and Figure 2.
Table 1. Selected Articles Related to Transfer Learning Models for Precision Medicine
Ref | Author | Year | Application Area | Dataset Source | Model Used | Transfer Method |
[31] | Amiri, et al. | 2024 | COVID-19 Decision Support System | COVID-19 World Dataset (Our World in Data), ECDC, GISAID | Neural Networks (Transfer Learning), Multi-attribute Decision-Making (MADM) | Fine-tuning (Transfer Learning) |
[32] | Haque, et al. | 2024 | Medical Imaging (Leukemia Diagnostics) | Kaggle Datasets (C-NMC, Leukemia Dataset 0.2) | AlexNet, Inception-ResNet, XceptionNet, RetinaNet, CenterNet, DCNN | Fine-tuning, Feature-based |
[33] | Mohan, et al. | 2024 | Medical Imaging (COVID-19 Detection from Chest X-rays) | COVID-19 Radiography Database (Kaggle), Chest X-ray Images Pneumonia and COVID-19 (Mendeley) | VGG16, Inception ResNet V2, CNN | Fine-tuning, Transfer Learning, CNN from Scratch |
[34] | Houssein, et al. | 2024 | Medical Imaging (Skin Cancer Classification) | HAM10000, ISIC-2019 | Deep Convolutional Neural Network (DCNN), VGG16, VGG19, DenseNet121, DenseNet201, MobileNetV2 | Fine-tuning, Transfer Learning |
[35] | Duan, et al. | 2024 | Medical Imaging (Meningioma Ki-67 Prediction) | MRI Images (318 cases) | Deep Transfer Learning (DTL), CNN (ResNet50) | Fine-tuning, Model-based |
[36] | Ma, et al. | 2024 | Medical Imaging (Autism Spectrum Disorder Classification) | Shenzhen Children's Hospital | Contrastive Variational AutoEncoder (CVAE), Random Forest | Transfer Learning, Fine-tuning |
[37] | Natha, et al. | 2024 | Medical Imaging (Brain Tumor Detection) | Kaggle Brain Tumor MRI Dataset | AlexNet, VGG19, Stack Ensemble Transfer Learning (SETL_BMRI) | Fine-tuning, Ensemble Learning |
[38] | Rasa, et al. | 2024 | Medical Imaging (Brain Tumor Detection) | BRATS 2015, Brain Tumor Classification Dataset (Kaggle) | VGG16, ResNet50, MobileNetV2, DenseNet201, EfficientNetB3, InceptionV3 | Fine-tuning |
[39] | Otaibi, et al. | 2024 | Medical Imaging (Brain Tumor Detection) | Multi-class Brain Tumor MRI Image Dataset (21,672 images) | 2D-CNN, VGG16, k-NN Classifier | Fine-tuning, Transfer learning |
[40] | Moran, et al. | 2024 | Medical Imaging (COPD Detection using ECG) | ECG signals (COPD and Healthy subjects) | Xception, VGG-19, InceptionResNetV2, DenseNet-121 | Fine-tuning and Transfer Learning |
[41] | Kumar, et al. | 2024 | Medical Imaging (Alzheimer's Disease Diagnosis) | Kaggle Dataset (12,936 MRI images) | GoogLeNet, FFNN | Fine-tuning, Feature extraction |
[42] | Azizian, et al. | 2024 | Genomics (miRNA-protein interactions) | RBPSuite, ENCODE, EVPsort, CLASH | Bi-LSTM, CNN, Cosine Similarity | Transfer learning, Cosine similarity |
[43] | Madduri, et al. | 2024 | Medical Imaging (Diabetic Eye Disease detection) | DRISHTI-GS, Messidor-2, Messidor, Kaggle cataract dataset | Modified ResNet-50, DenseUNet | Two-phase transfer learning (ResNet-50 for classification, DenseUNet for segmentation) |
[44] | Gore, et al. | 2024 | Disease Classification (Non-communicable diseases) | NCBI GEO (GSE datasets) | Variational Autoencoder (VAE) with transfer learning from CancerNet | Transfer learning (CancerNet to NCDs) |
[45] | Taghizadeh, et al. | 2024 | EEG Motor Imagery Classification (Brain-Computer Interface) | Physionet MI Dataset | 1D-CNN with Semi-deep Fine-tuning | Transfer learning with feature-extracted data, fine-tuning pre-trained model |
[46] | Raza, et al. | 2024 | Medical Imaging (Diabetic Retinopathy, MRI Brain Tumor) | Diabetic Retinopathy Dataset, MRI Brain Tumor Dataset | Mobile-Net, CNN, PSO-Optimized | Transfer learning with Particle Swarm Optimization (PSO) and Constriction Factor |
[47] | Ansari, et al. | 2024 | Medical Imaging (Lung Cancer Detection) | LIDC-IDRI | ResNet-50, VGG-16, ResNet-101, VGG-19, DenseNet-201, EfficientNet-B4 | Transfer learning with hyperparameter tuning |
[48] | Alturki, et al. | 2024 | Clinical Data (Chronic Kidney Disease Prediction) | UCI CKD dataset | XGBoost, Random Forest, Extra Trees Classifier (TrioNet ensemble) | KNN imputer for missing values, SMOTE for class imbalance |
[49] | Jakkaladiki, et al. | 2024 | Medical Imaging (Breast Cancer Diagnosis) | BreakHis (Kaggle), Wisconsin Breast Cancer (UCI) | CNN, DenseNet, Hybrid Transfer Learning | Transfer learning with attention mechanism |
[50] | Ragab, et al. | 2024 | Medical Imaging (COVID-19 detection from chest X-rays) | COVID-ChestXRay Dataset | DenseNet121, Autoencoder-LSTM, Firefly Algorithm (FFA) | Hybrid transfer learning with FFA for hyperparameter optimization |
[51] | Koshy, et al. | 2024 | Medical Imaging (Breast Cancer Histopathology Classification) | BreaKHis | ResNet-18, CNN, Levenberg–Marquardt Optimization | Transfer learning with fine-tuning |
[52] | Salinas, et al. | 2024 | Emotion Recognition (Driver Emotion) | CARLA Simulator (Simulated Driving Data) | CNN (VGG16, Inception V3, EfficientNet) | Transfer learning with fine-tuning |
[53] | Li, et al. | 2024 | Clinical Data (Pediatric Knowledge Extraction) | Mass General Brigham (MGB), Boston Children’s Hospital (BCH) | MUGS (Multisource Graph Synthesis), SVD | Transfer learning, Graph-based feature engineering |
[54] | Wang, et al. | 2024 | Clinical Data (Epilepsy Recognition) | University of Bonn EEG Dataset | Multi-View Transfer Learning (MVTL-LSR), CNN | Multi-view & transfer learning with privacy protection |
[55] | Enguita, et al. | 2024 | Genomics (DNA Methylation) | EWAS Data Hub (Illumina 450K and EPIC arrays) | Autoencoders (NCAE), Deep Neural Networks (DNN) | Transfer learning, NCAE embedding |
[56] | Roy, et al. | 2024 | Clinical Data (Human Activity Detection) | WISDM (Wireless Sensor Data Mining Lab dataset) | Semi-Supervised Learning (SSL), k-means, GMM | On-device learning with sparse labeling and clustering |
[57] | Ragab, et al. | 2024 | Medical Imaging (Colorectal Cancer Detection) | Warwick-QU Dataset | Dense-EfficientNet, Slime Mould Algorithm (SMA), Deep Hopfield Neural Network (DHNN) | Transfer learning, hyperparameter optimization |
[58] | Hoseny, et al. | 2024 | Medical Imaging (Diabetic Retinopathy Classification) | Kaggle Dataset (Diabetic Retinopathy Images) | VGG16, CNN, AE, CLAHE | Transfer learning with data cleansing and enhancement filters |
[59] | Siddique, et al. | 2024 | Medical Imaging (Tumor Classification) | Kaggle (Brain Tumor Images) | Inception-V3, CNN | Transfer learning with Particle Swarm Optimization (PSO) |
[60] | Xue, et al. | 2024 | Clinical Data (Parkinson’s Disease Severity Prediction) | Parkinson's Telemonitoring Dataset | Random Forest (RF), Shapley Value, Game-based Transfer | Patient-specific Game-based Transfer (PSGT), Instance Transfer |
[61] | Kunjumon, et al. | 2024 | Medical Imaging (Esophageal Cancer Diagnosis) | Kaggle Endoscopic Image Dataset | Inception-ResNet V2, CNN | Transfer learning with fine-tuning |
[62] | Ajani, et al. | 2024 | Medical Imaging (COVID-19 Screening) | COVID-19 Chest X-ray (CXR), CT Dataset | GoogleNet, SqueezeNet, CNN | Transfer learning with fine-tuning |
[63] | Djaroudib, et al. | 2024 | Medical Imaging (Skin Cancer Diagnosis) | Kaggle (HAM10000, Skin Cancer MNIST) | VGG16, CNN | Transfer learning with fine-tuning |
[64] | Alzubaidi, et al. | 2024 | Medical Imaging (Shoulder Implant Classification) | UCI Shoulder Implant X-ray Dataset | Xception, InceptionResNetV2, MobileNetV2, EfficientNet, DarkNet19 | Self-Supervised Pertaining (SSP), Transfer learning |
[65] | Hong, et al. | 2024 | Clinical Data (Risk Prediction) | FOS, ARIC, MESA, REGARDS | Logistic Regression, Translasso | Federated learning, Transfer learning, ROSE |
[66] | Alnuaimi, et al. | 2024 | Medical Imaging (Skin Diseases Detection) | Kaggle DermNet, Google Images, Atlas Dermatology | MobileNet, DenseNet121, CNN | Transfer learning with fine-tuning |
[67] | Xiang, et al. | 2024 | Clinical Data (Predictive Modeling) | PM2.5, IHS, HUA, Wine, eICU | Random Forest (RF), Federated Learning (FL), Model Averaging | Federated Transfer Learning (FTRF) |
[68] | Khouadja, et al. | 2024 | Medical Imaging (Lung Cancer Diagnosis) | Military Hospital of Tunis (DICOM CT scans) | ResNet50, InceptionV3, VGG16 | Transfer learning with pre-trained 3D ResNet models from Tencent MedicalNet |
[69] | Sambyal, et al. | 2024 | Medical Imaging (Calibration of Deep Neural Networks) | Diabetic Retinopathy, Histopathologic Cancer, COVID-19 datasets | ResNet18, ResNet50, WideResNet | Transfer learning, Rotation-based self-supervised learning (SSL) |
[70] | Benbakreti, et al. | 2024 | Medical Imaging (Breast Cancer Classification) | Inbreast, MIAS, DDSM | ResNet18, AlexNet, InceptionV3 | Transfer learning with pre-trained models |
The section presents a comprehensive review of how Transfer Learning (TL) models are currently being applied across various fields in precision medicine. These models are crucial for advancing medical diagnostics, particularly in contexts where data scarcity or complexity poses significant challenges. One of the key areas of medical imaging is the application of TL for diagnostic purposes, such as COVID-19 detection and cancer classification. Amiri et al. [31] developed a COVID-19 decision support system using neural networks, leveraging datasets from sources like Our World in Data and GISAID, with a fine-tuning method. Similarly, Ragab et al. [50] focused on chest X-ray images for COVID-19 detection using DenseNet121 and Autoencoder-LSTM with hybrid transfer learning for hyperparameter optimization. This shows the broad utility of TL in diagnosing infectious diseases in urgent public health situations. Additionally, for skin cancer classification, Houssein et al. [34] applied deep convolutional neural networks (DCNN) and VGG models, showcasing the effectiveness of TL in dermatological diagnostics using datasets like HAM10000 and ISIC-2019. TL models are also widely used for brain tumor detection and leukemia diagnostics. Natha et al. [37] and Rasa et al. [38] explored TL for brain tumor detection, using models such as ResNet50, VGG16, and MobileNetV2 on MRI datasets from Kaggle and BRATS 2015. Their results underscore the potential of TL to enhance tumor detection and improve diagnostic accuracy. For leukemia detection, Haque et al. [32] employed fine-tuning and feature-based TL methods using AlexNet and Inception-ResNet on datasets from Kaggle, demonstrating how TL helps in processing complex medical imaging data for blood cancer diagnoses. In the field of genomics, Azizian et al. [42] utilized TL to study miRNA-protein interactions, applying Bi-LSTM and CNN models with cosine similarity methods. This approach facilitates the analysis of genomic data, which often suffers from sparsity. Similarly, in chronic kidney disease prediction, Alturki et al. [48] used XGBoost and random forest classifiers to make predictions based on clinical datasets, demonstrating how TL is applied to clinical data to forecast disease outcomes.
Clinical data is another area where TL has shown significant promise, particularly in predictive modeling. Wang et al. [55] employed federated learning combined with TL to analyze risk factors for chronic diseases using datasets like FOS, ARIC, and MESA, ensuring data privacy while still enhancing predictive accuracy. Xue et al. [60] applied TL with Random Forest and Shapley value models to predict Parkinson’s disease severity using telemonitoring datasets, illustrating the integration of TL in clinical decision-making processes. In diabetic retinopathy classification, Hoseny et al. [58] used transfer learning with fine-tuning on VGG16 and CNN models, focusing on the Kaggle diabetic retinopathy dataset. Their research shows how TL helps in processing and enhancing medical images for ophthalmic disease diagnosis. Similarly, in breast cancer diagnosis, Jakkaladiki et al. [49] used hybrid TL models combined with attention mechanisms, demonstrating how TL can aid in cancer detection from imaging datasets like BreakHis and Wisconsin Breast Cancer. Several other studies also demonstrate TL applications in disease classification, emotion recognition, and activity detection. For example, Kunjumon et al. [61] applied transfer learning with fine-tuning to diagnose esophageal cancer using endoscopic image datasets. Similarly, Ajani et al. [62] focused on COVID-19 screening from chest X-rays and CT images using GoogleNet and SqueezeNet, further emphasizing the role of TL in improving diagnostic tools for infectious diseases. Lastly, federated learning is becoming increasingly important in medical applications as demonstrated by Xiang et al. [67], who applied federated transfer learning for predictive modeling in clinical data, further enhancing the utility of TL across decentralized datasets while maintaining privacy.

Figure 2. Applications of Transfer Learning Models in Precision Medicine
- STRENGTHS AND LIMITATIONS
- Strengths of Transfer Learning in Precision Medicine
Transfer learning (TL) offers significant advantages in precision medicine, particularly in addressing the constraints arising from scarce medical datasets. One of its key advantages is its capacity to exploit pretrained models built on large, heterogeneous datasets, which can be further tuned or customized to particular clinical applications. This approach lessens the requirement for large amounts of labeled data in target domains, which is often difficult and expensive to obtain in healthcare contexts, such as in rare diseases or specialized imaging datasets [71][72]. By transferring knowledge from a data-rich source domain to a data-limited target domain, TL enables the development of highly accurate models without requiring extensive data collection for every medical task. Another strength of TL in precision medicine is its ability to enhance the generalizability of machine learning models. Fine-tuning models on domain-specific datasets helps improve performance on small datasets while reducing overfitting. This is particularly valuable in medical applications like diagnostic imaging or genomics, where variability in patient data can significantly impact the model's accuracy. For example, TL models in medical imaging, for example, those applied to brain tumor detection or skin cancer classification, have shown impressive results when fine-tuned from pretrained architectures such as VGG16, ResNet, and DenseNet [73][74]. Furthermore, TL models can be adapted to a broad array of tasks, including disease detection, prediction, and classification, across different medical domains, enhancing their versatility in clinical settings [75][76].
- Limitations of Transfer Learning in Precision Medicine
Despite its many strengths, TL in precision medicine also faces a number of constraints. One of the primary challenges is the risk of negative transfer, where the knowledge transferred from the source domain may not be relevant or beneficial for the target domain. This problem emerges when the differences between the source and target domains are too large, leading to suboptimal model effectiveness. As an example, algorithms developed on general medical imaging datasets may struggle to perform well when applied to highly specialized conditions or rare diseases with minimal data availability [77][78]. Additionally, TL models are often sensitive to domain shifts, such as variations in imaging equipment or demographic characteristics, which can compromise their ability to generalize effectively in real-world clinical settings. Another limitation is the dependency upon the standard of the pre-trained models along with the originating datasets. The success of TL heavily relies on the quality and representativeness of the dataset used for the model’s initial training. If the source data is biased, unbalanced, or unrepresentative of the target population, the transfer of knowledge may lead to poor model performance or biased predictions in the target domain. As an illustration, TL models built on data originating from a particular geographic location or patient group might fail to achieve satisfactory results when implemented on groups with distinct demographic traits or varying medical conditions [79][80]. Furthermore, while TL can diminish the dependence on vast quantities of annotated datasets, the task of refining models still demands substantial computing capacity and specialized knowledge, making it less accessible for some healthcare institutions, especially those with limited technical infrastructure [81].
- EMERGING TRENDS AND RESEARCH GAPS
The application of the concept of transfer learning (TL) in precision medicine has attracted notable momentum over the past few years, driven by the increasing need for high-quality models in medical domains that often suffer from limited labeled data. One of the prominent trends is the integration of TL with advanced deep learning frameworks, especially CNNs, in medical imaging and genomics. For instance, TL methods are being used to adapt models trained using extensive datasets like ImageNet or GoogleNet to specialized medical datasets, which enables high performance even with relatively small medical image datasets. This approach has been particularly beneficial in areas like cancer detection, skin lesion classification, and brain tumor detection, where acquiring large annotated datasets is costly and time-consuming. Additionally, fine-tuning pre-trained models has become a common practice, significantly improving model accuracy without requiring the exhaustive retraining of models from scratch. Another emerging trend is the increasing use of multi-modal transfer learning, where models leverage information originating from multiple sources, including medical imaging, clinical documentation, and genomic data. This hybrid approach allows for a more comprehensive analysis of patient data, integrating insights from multiple domains to generate a broader and more integrated perspective on patient health. For example, in the detection of diabetic retinopathy, researchers combine image data from retinal scans with patient health records to create more robust predictive models. The combination of clinical data and medical imaging, aided by TL, has the potential to provide more accurate disease predictions, as demonstrated in disorders like Alzheimer’s disease and Parkinson’s disease. Furthermore, the adoption of ensemble learning, where multiple pre-trained models are combined, is helping to enhance performance by leveraging the strengths of different architectures.
Despite these advancements, several research gaps persist. One significant challenge in TL for precision medicine is the issue of negative transfer, where the transmission of knowledge from the origin domain to the destination domain does not improve, or even degrades, model performance. This is especially problematic when the allocation of data between the source and destination domains is quite different, leading to suboptimal results. Another gap is related to issues of data bias and confidentiality, especially within the framework of sensitive healthcare information. There is an increasing need for techniques that mitigate biases in medical datasets, such as those related to race, gender, or geographical location, to guarantee that models achieve broad applicability and equity across varied populations. Additionally, while data privacy remains a critical concern, federated learning models, where data remains decentralized, are showing promise in addressing these issues while still allowing for collaborative learning across multiple institutions. Lastly, the complexity of integrating TL into clinical settings remains a substantial challenge. Despite the promise of TL in research, translating these models into real-world clinical applications requires addressing barriers such as the lack of standardized datasets, variability in healthcare systems, and the interpretability of AI models. Clinicians often require clear, understandable insights from models to make informed decisions, which calls for greater efforts in making AI models more explainable. As the field moves forward, the integration of explainable AI (XAI) with transfer learning techniques in precision medicine is an area that warrants significant research, particularly in ensuring that medical professionals can trust and interpret AI-based recommendations effectively.
- CONCLUSIONS
The integration of Transfer Learning (TL) in precision medicine has shown considerable promise across various medical domains, offering an effective solution to address the challenges associated with restricted access to data and high computational costs. As demonstrated by the numerous studies presented in the table, TL has been effectively implemented across multiple fields, such as medical imaging, genomics, clinical data analysis, disease classification, and emotion recognition. The ability of TL models to leverage pre-trained networks and customize them for data tailored to particular tasks has led to improved model effectiveness in applications spanning from early disease detection to patient-specific predictions. Within the domain of medical imaging, TL has played a crucial role in improving the accuracy and efficiency of diagnostic systems, particularly in the detection of diseases like cancer, COVID-19, and brain tumors. By using models such as CNNs, ResNet, and DenseNet, TL has been able to improve feature extraction and decision-making processes in imaging modalities like X-rays, MRIs, and CT scans. These models, when fine-tuned with domain-specific data, have demonstrated significant performance gains, enabling more precise and rapid diagnosis even in resource-constrained environments. In genomics and clinical data analysis, TL has facilitated the extraction of meaningful patterns from complex datasets, such as DNA methylation and pediatric health data. Techniques like Bi-LSTM and Random Forest, integrated with TL approaches, have provided novel insights into disease mechanisms and improved predictive models for conditions like chronic kidney disease and Parkinson’s disease. The transferability of knowledge from general domains to specialized medical contexts has proven to be a game-changer, especially in rare diseases where data scarcity is a major challenge. Moreover, TL has made significant contributions to disease classification and emotion recognition. In the domain of chronic disease prediction and classification, TL models have achieved high accuracy in predicting the onset and progression of diseases like lung cancer and diabetes. Similarly, in emotion recognition, TL has been employed to analyze complex datasets, such as driving simulation data, to understand human behavior better and predict outcomes in real-world scenarios.
DECLARATION
Author Contribution
All authors contributed equally to the main contributor to this paper. All authors read and approved the final paper.
Acknowledgement
The authors would like to acknowledge the Department of Medical Technology, Institut Teknologi Sepuluh Nopember, for the facilities and support in this research. The authors also gratefully acknowledge financial support from the Institut Teknologi Sepuluh Nopember for this work, under project scheme of the Publication Writing and IPR Incentive Program (PPHKI) 2025.
Conflicts of Interest
The authors declare no conflict of interest.
REFERENCES
- M. Edvardsson and M. K. Heenkenda, “Precision Medicine: Personalizing Healthcare by Bridging Aging, Genetics, and Global Diversity,” Healthcare, vol. 13, no. 13, pp. 1529–1529, 2025, https://doi.org/10.3390/healthcare13131529.
- L. R. Moffitt, N. Karimnia, A. L. Wilson, A. N. Stephens, G.-Y. Ho, and M. Bilandzic, “Challenges in Implementing Comprehensive Precision Medicine Screening for Ovarian Cancer,” Current Oncology, vol. 31, no. 12, pp. 8023–8038, 2024, https://doi.org/10.3390/curroncol31120592.
- M. Hassan, F. M. Awan, A. Naz, E. J. d. A.-Galiana, O. Alvarez, A. Cernea, L. F.-Brillet, J. L. F.-Martínez, and A. Kloczkowski, “Innovations in Genomics and Big Data Analytics for Personalized Medicine and Health Care: A Review,” International Journal of Molecular Sciences, vol. 23, no. 9, p. 4645, 2022, https://doi.org/10.3390/ijms23094645.
- S. M. Varnosfaderani and M. Forouzanfar, “The Role of AI in Hospitals and Clinics: Transforming Healthcare in the 21st Century,” Bioengineering, vol. 11, no. 4, pp. 1–38, 2024, https://doi.org/10.3390/bioengineering11040337
- A. Farmanesh, R. G. Sanchis, and J. Ordieres-Meré, “Comparison of Deep Transfer Learning Against Contrastive Learning in Industrial Quality Applications for Heavily Unbalanced Data Scenarios When Data Augmentation Is Limited,” Sensors, vol. 25, no. 10, p. 3048, 2025, https://doi.org/10.3390/s25103048.
- Y. Qiu, F. Lin, W. Chen, and M. Xu, “Pre-training in Medical Data: A Survey,” Machine Intelligence Research, vol. 20, no. 2, pp. 147-179 2023, https://doi.org/10.1007/s11633-022-1382-8.
- M.-S. Șerbănescu, L. Streba, A. D. Demetrian, A.-G. Gheorghe, M. Mămuleanu, D.-N. Pirici, and C.-T. Streba, “Transfer Learning-Based Integration of Dual Imaging Modalities for Enhanced Classification Accuracy in Confocal Laser Endomicroscopy of Lung Cancer,” Cancers, vol. 17, no. 4, pp. 611–611, 2025, https://doi.org/10.3390/cancers17040611.
- D. Du, F. Zhong, and L. Liu, “Enhancing recognition and interpretation of functional phenotypic sequences through fine-tuning pre-trained genomic models,” Journal of Translational Medicine, vol. 22, no. 1, 2024, https://doi.org/10.1186/s12967-024-05567-z.
- S. K. Zhou, H. Greenspan, C. Davatzikos, J. S. Duncan, B. v. Ginneken, A. Madabhushi, J. L. Prince, D. Rueckert, and R. M. Summers, “A Review of Deep Learning in Medical Imaging: Imaging Traits, Technology Trends, Case Studies With Progress Highlights, and Future Promises,” Proceedings of the IEEE, vol. 109, no. 5, pp. 1–19, 2021, https://doi.org/10.1109/jproc.2021.3054390.
- S. Mohamedikbal, H. A. Al-Mamun, M. S. Bestry, J. Batley, and D. Edwards, “Integrating multi-omics and machine learning for disease resistance prediction in legumes,” Theoretical and Applied Genetics, vol. 138, no. 7, 2025, https://doi.org/10.1007/s00122-025-04948-2.
- Z. Zhao, L. Alzubaidi, J. Zhang, Y. Duan, and Y. Gu, “A comparison review of transfer learning and self-supervised learning: Definitions, applications, advantages and limitations,” Expert Systems with Applications, vol. 242, p. 122807, 2024, https://doi.org/10.1016/j.eswa.2023.122807.
- T. Islam, M. S. Hafiz, J. R. Jim, M. M. Kabir, and M. F. Mridha, “A systematic review of deep learning data augmentation in medical imaging: Recent advances and future research directions,” Healthcare Analytics, vol. 5, pp. 100340–100340, 2024, https://doi.org/10.1016/j.health.2024.100340.
- A. Rahujo, D. Atif, S. A. Inam, A. A. Khan, and S. Ullah, “A survey on the applications of transfer learning to enhance the performance of large language models in healthcare systems,” Discover Artificial Intelligence, vol. 5, no. 1, 2025, https://doi.org/10.1007/s44163-025-00339-0.
- G. Ayana, K. Dese, A. M. Abagaro, K. C. Jeong, S.-D. Yoon, and S. Choe, “Multistage transfer learning for medical images,” Artificial Intelligence Review, vol. 57, no. 9, 2024, https://doi.org/10.1007/s10462-024-10855-7.
- S. Rajendran, W. Pan, M. R. Sabuncu, Y. Chen, J. Zhou, and F. Wang, “Learning across diverse biomedical data modalities and cohorts: Challenges and opportunities for innovation,” Patterns, vol. 5, no. 2, pp. 100913–100913, 2024, https://doi.org/10.1016/j.patter.2023.100913.
- K. A.-Moura, L. Souza, T. A. d. Oliveira, M. S. Rocha, A. C. F. D. Moraes, and A. C. Filho, “Prediction of Hypertension in the Pediatric Population Using Machine Learning and Transfer Learning: A Multicentric Analysis of the SAYCARE Study,” International Journal of Public Health, vol. 70, 2025, https://doi.org/10.3389/ijph.2025.1607944.
- V. Mehan, “Advanced artificial intelligence driven framework for lung cancer diagnosis leveraging SqueezeNet with machine learning algorithms using transfer learning,” Medicine in Novel Technology and Devices, vol. 27, p. 100383, 2025, https://doi.org/10.1016/j.medntd.2025.100383.
- S. Bashath, N. Perera, S. Tripathi, K. Manjang, M. Dehmer, and F. E. Streib, “A data-centric review of deep transfer learning with applications to text data,” Information Sciences, vol. 585, pp. 498–528, 2022, https://doi.org/10.1016/j.ins.2021.11.061.
- M. Jafari, X. Tao, P. Barua, R.-S. Tan, and U. R. Acharya, “Application of Transfer Learning for Biomedical Signals: A Comprehensive Review of the Last Decade (2014-2024),” Information Fusion, vol. 118, pp. 102982–102982, 2025, https://doi.org/10.1016/j.inffus.2025.102982.
- X. Zhang, J. Wang, J. Wei, X. Yuan, and M. Wu, “A Review of Non-Fully Supervised Deep Learning for Medical Image Segmentation,” Information, vol. 16, no. 6, pp. 433–433, 2025, https://doi.org/10.3390/info16060433.
- N. Erfanian, A. A. Heydari, A. M. Feriz, P. Iañez, A. Derakhshani, M. Ghasemigol, M. Farahpour, S. M. Razavi, S. Nasseri, H. Safarpour, and A. Sahebkar, “Deep learning applications in single-cell genomics and transcriptomics data analysis,” Biomedicine & Pharmacotherapy, vol. 165, p. 115077, 2023, https://doi.org/10.1016/j.biopha.2023.115077.
- L. Chato and E. Regentova, “Survey of Transfer Learning Approaches in the Machine Learning of Digital Health Sensing Data,” Journal of Personalized Medicine, vol. 13, no. 12, pp. 1703–1703, 2023, https://doi.org/10.3390/jpm13121703.
- P. Li, “A Multi-scale Attention-Based Transfer Model for Cross-bearing Fault Diagnosis,” International Journal of Computational Intelligence Systems, vol. 17, no. 1, 2024, https://doi.org/10.1007/s44196-024-00414-x.
- J. Meng, H. Lin, and Y. Li, “Knowledge transfer based on feature representation mapping for text classification,” Expert Systems with Applications, vol. 38, no. 8, pp. 10562–10567, 2011, https://doi.org/10.1016/j.eswa.2011.02.085.
- M. Wang, Q. Song, and W. Lai, “On Model-Based Transfer Learning Method for the Detection of Inter-Turn Short Circuit Faults in PMSM,” Sensors, vol. 23, no. 22, pp. 9145–9145, 2023, https://doi.org/10.3390/s23229145.
- K. A. A. Mahmoud, M. M. Badr, N. A. Elmalhy, R. A. Hamdy, S. Ahmed, and A. A. Mordi, “Transfer learning by fine-tuning pre-trained convolutional neural network architectures for switchgear fault detection using thermal imaging,” Alexandria Engineering Journal, vol. 103, pp. 327–342, 2024, https://doi.org/10.1016/j.aej.2024.05.102.
- H. E. Kim, A. C.-Linan, N. Santhanam, M. Jannesari, M. E. Maros, and T. Ganslandt, “Transfer learning for medical image classification: a literature review,” BMC Medical Imaging, vol. 22, no. 1, 2022, https://doi.org/10.1186/s12880-022-00793-7.
- A. W. Salehi, S. Khan, G. Gupta, B. I. Alabduallah, A. Almjally, H. Alsolai, T. Siddiqui, and A. Mellit, “A Study of CNN and Transfer Learning in Medical Imaging: Advantages, Challenges, Future Scope,” Sustainability, vol. 15, no. 7, p. 5930, 2023, https://doi.org/10.3390/su15075930.
- Y.-M. Chen, T.-H. Hsiao, C.-H. Lin, and Y. C. Fann, “Unlocking precision medicine: clinical applications of integrating health records, genetics, and immunology through artificial intelligence,” Journal of Biomedical Science, vol. 32, no. 1, 2025, https://doi.org/10.1186/s12929-024-01110-w.
- R. Aswini, R. Angelin Rajan, B. Saranya, and K. Elumalai, “AI‐Enhanced Predictive Imaging in Precision Medicine: Advancing Diagnostic Accuracy and Personalized Treatment,” iRADIOLOGY, vol. 3, no. 4, pp. 261-278 2025, https://doi.org/10.1002/ird3.70027.
- A. S. Amiri, A. Babaei, V. Simic, and E. B. Tirkolaee, “A variant-informed decision support system for tackling COVID-19: a transfer learning and multi-attribute decision-making approach,” PeerJ Computer Science, vol. 10, p. e2321, 2024, https://doi.org/10.7717/peerj-cs.2321.
- R. Haque et al., “Advancing Early Leukemia Diagnostics: A Comprehensive Study Incorporating Image Processing and Transfer Learning,” BioMedInformatics, vol. 4, no. 2, pp. 966–991, 2024, https://doi.org/10.3390/biomedinformatics4020054.
- G. Mohan, M. M. Subashini, S. Balan, and S. Singh, “A multiclass deep learning algorithm for healthy lung, Covid-19 and pneumonia disease detection from chest X-ray images,” Discover Artificial Intelligence, vol. 4, no. 1, 2024, https://doi.org/10.1007/s44163-024-00110-x.
- E. H. Houssein, D. A. Abdelkareem, G. Hu, M. A. Hameed, I. A. Ibrahim, and M. Younan, “An effective multiclass skin cancer classification approach based on deep convolutional neural network,” Cluster computing, vol. 27, no. 9, pp. 12799-12819 2024, https://doi.org/10.1007/s10586-024-04540-1.
- C. Duan, D. Hao, J. Cui, G. Wang, W. Xu, N. Li, and X. Liu, “An MRI-Based Deep Transfer Learning Radiomics Nomogram to Predict Ki-67 Proliferation Index of Meningioma,” Journal of Imaging Informatics in Medicine, vol. 37, no. 2, pp. 510–519, 2024, https://doi.org/10.1007/s10278-023-00937-3.
- R. Ma, R. Xie, Y. Wang, J. Meng, Y. Wei, Y. Cai, W. Xi, and Y. Pan, “Autism Spectrum Disorder Classification with Interpretability in Children Based on Structural MRI Features Extracted Using Contrastive Variational Autoencoder,” Big Data Mining and Analytics, vol. 7, no. 3, pp. 781–793, 2024, https://doi.org/10.26599/bdma.2024.9020004.
- S. Natha, U. Laila, I. A. Gashim, K. Mahboob, M. N. Saeed, and K. M. Noaman, “Automated Brain Tumor Identification in Biomedical Radiology Images: A Multi-Model Ensemble Deep Learning Approach,” Applied Sciences, vol. 14, no. 5, p. 2210, 2024, https://doi.org/10.3390/app14052210.
- S. M. Rasa, M. M. Islam, M. A. Talukder, M. A. Uddin, M. Khalid, M. Kazi, and M. Z. Kazi, “Brain tumor classification using fine-tuned transfer learning models on magnetic resonance imaging (MRI) images,” Digital Health, vol. 10, 2024, https://doi.org/10.1177/20552076241286140.
- S. A.-Otaibi, A. Rehman, A. Raza, J. Alyami, and T. Saba, “CVG-Net: novel transfer learning based deep features for diagnosis of brain tumors using MRI scans,” PeerJ Computer Science, vol. 10, p. e2008, 2024, https://doi.org/10.7717/peerj-cs.2008.
- I. Moran, D. T. Altılar, M. K. Uçar, C. Bilgin, and M. R. Bozkurt, “Deep Transfer Learning for Chronic Obstructive Pulmonary Disease Detection Utilizing Electrocardiogram Signals,” IEEE Access, vol. 11, pp. 40629–40644, 2023, https://doi.org/10.1109/access.2023.3269397.
- P. N. Kumar and L. P. Maguluri, “Deep Learning-based Classification of MRI Images for Early Detection and Staging of Alzheimer’s Disease,” International Journal of Advanced Computer Science and Applications, vol. 15, no. 5, 2024, https://doi.org/10.14569/ijacsa.2024.0150545.
- S. Azizian and J. Cui, “DeepMiRBP: a hybrid model for predicting microRNA-protein interactions based on transfer learning and cosine similarity,” BMC Bioinformatics, vol. 25, no. 1, 2024, https://doi.org/10.1186/s12859-024-05985-2.
- V. K. Madduri and B. S. Rao, “Detection and diagnosis of diabetic eye diseases using two phase transfer learning approach,” PeerJ Computer Science, vol. 10, pp. e2135–e2135, 2024, https://doi.org/10.7717/peerj-cs.2135.
- S. Gore, B. Meche, D. Shao, B. Ginnett, K. Zhou, and R. K. Azad, “DiseaseNet: a transfer learning approach to noncommunicable disease classification,” BMC Bioinformatics, vol. 25, no. 1, 2024, https://doi.org/10.1186/s12859-024-05734-5.
- M. Taghizadeh, F. Vaez, and M. Faezipour, “EEG Motor Imagery Classification by Feature Extracted Deep 1D-CNN and Semi-Deep Fine-Tuning,” IEEE Access, vol. 12, pp. 111265–111279, 2024, https://doi.org/10.1109/access.2024.3430838.
- A. Raza, S. Musa, A. Shahrafidz, M. M. Alam, M. M. Su’ud, and D. F. Noor, “Enhancing Medical Image Classification through PSO-Optimized Dual Deterministic Approach and Robust Transfer Learning,” IEEE Access, pp. 1–1, 2024, https://doi.org/10.1109/access.2024.3504266.
- M. M. Ansari, S. Kumar, U. Tariq, M. B. Heyat, F. Akhtar, M. A. Hayat, E. Sayeed, S. Parveen, and D. Pomary, “Evaluating CNN Architectures and Hyperparameter Tuning for Enhanced Lung Cancer Detection Using Transfer Learning,” Journal of Electrical and Computer Engineering, vol. 2024, no. 1, 2024, https://doi.org/10.1155/2024/3790617.
- N. Alturki, A. Altamimi, M. Umer, O. Saidani, A. Alshardan, S. Alsubai, M. Omar, and I. Ashraf, “Improving Prediction of Chronic Kidney Disease Using KNN Imputed SMOTE Features and TrioNet Model,” Computer Modeling in Engineering & Sciences, vol. 0, no. 0, pp. 1–10, 2023, https://doi.org/10.32604/cmes.2023.045868.
- S. P. Jakkaladiki and F. Maly, “Integrating hybrid transfer learning with attention-enhanced deep learning models to improve breast cancer diagnosis,” PeerJ Computer Science, vol. 10, p. e1850, 2024, https://doi.org/10.7717/peerj-cs.1850.
- M. Ragab, M. W. A.-Rabia, S. S. Binyamin, and A. A. Aldarmahi, “Intelligent Firefly Algorithm Deep Transfer Learning Based COVID-19 Monitoring System,” Computers, materials & continua/Computers, materials & continua (Print), vol. 74, no. 2, pp. 2889–2903, 2022, https://doi.org/10.32604/cmc.2023.032192.
- S. S. Koshy and L. J. Anbarasi, “LMHistNet: Levenberg–Marquardt Based Deep Neural Network for Classification of Breast Cancer Histopathological Images,” IEEE access, pp. 1–1, 2024, https://doi.org/10.1109/access.2024.3385011.
- C. H. E.-Salinas, H. L.-García, J. M. C.-Padilla, C. B.-Huidobro, N. K. G. Rosales, D. Rondon, and K. O. V.-Condori, “Multimodal driver emotion recognition using motor activity and facial expressions,” Frontiers in Artificial Intelligence, vol. 7, 2024, https://doi.org/10.3389/frai.2024.1467051.
- M. Li, X. Li, K. Pan, A. Geva, D. Yang, S. M. Sweet, C.-L. Bonzel, V. A. Panickan, X. Xiong, K. Mandl, and T. Cai, “Multisource representation learning for pediatric knowledge extraction from electronic health records,” npj Digital Medicine, vol. 7, no. 1, 2024, https://doi.org/10.1038/s41746-024-01320-4.
- J. Wang, B. Li, C. Qiu, X. Zhang, Y. Cheng, P. Wang, T. Zhou, H. Ge, Y. Zhang, and J. Cai, “Multi-View & Transfer Learning for Epilepsy Recognition Based on EEG Signals,” Computers, Materials & Continua, vol. 75, no. 3, pp. 4843–4866, 2023, https://doi.org/10.32604/cmc.2023.037457.
- D. M.-Enguita, S. K. Dwivedi, R. Jörnsten, and M. Gustafsson, “NCAE: data-driven representations using a deep network-coherent DNA methylation autoencoder identify robust disease and risk factor signatures,” Briefings in Bioinformatics, vol. 24, no. 5, p. bbad293, 2023, https://doi.org/10.1093/bib/bbad293.
- A. Roy, H. Dutta, A. K. Bhuyan, and S. Biswas, “On-Device Semi-Supervised Activity Detection: A New Privacy-Aware Personalized Health Monitoring Approach,” Sensors, vol. 24, no. 14, p. 4444, 2024, https://doi.org/10.3390/s24144444.
- M. Ragab, M. M. Mahmoud, A. H. Asseri, H. Choudhry, and H. A. Yacoub, “Optimal Deep Transfer Learning Based Colorectal Cancer Detection and Classification Model,” Computers, Materials & Continua, vol. 74, no. 2, pp. 3279–3295, 2023, https://doi.org/10.32604/cmc.2023.031037.
- H. M. E.-Hoseny, H. F. Elsepae, W. A. Mohamed, and A. S. Selmy, “Optimized Deep Learning Approach for Efficient Diabetic Retinopathy Classification Combining VGG16-CNN,” Computers, materials & continua/Computers, materials & continua (Print), vol. 77, no. 2, pp. 1855–1872, 2023, https://doi.org/10.32604/cmc.2023.042107.
- A. A. Siddique, A. Raza, M. S. Alshehri, N. Alasbali, and S. F. Abbasi, “Optimizing Tumor Classification Through Transfer Learning and Particle Swarm Optimization-Driven Feature Extraction,” IEEE Access, vol. 12, pp. 85929–85939, 2024, https://doi.org/10.1109/access.2024.3412412.
- Z. Xue, H. Lu, T. Zhang, and M. A. Little, “Patient-specific game-based transfer method for Parkinson’s disease severity prediction,” Artificial Intelligence in Medicine, pp. 102810–102810, 2024, https://doi.org/10.1016/j.artmed.2024.102810.
- S. P. Kunjumon and S. F. Stephen, “Revolutionizing Esophageal Cancer Diagnosis: A Deep Learning-Based Method in Endoscopic Images,” International Journal of Advanced Computer Science and Applications, vol. 15, no. 7, 2024, https://doi.org/10.14569/ijacsa.2024.0150728.
- O. S. Ajani, H. Obasekore, B.-Y. Kang, and M. Rammohan, “Robotic Assistance in Radiology: A Covid-19 Scenario,” IEEE Access, vol. 11, pp. 49785–49793, 2023, https://doi.org/10.1109/access.2023.3277526.
- K. Djaroudib, P. Lorenz, R. B. Bouzida, and H. Merzougui, “Skin Cancer Diagnosis Using VGG16 and Transfer Learning: Analyzing the Effects of Data Quality over Quantity on Model Efficiency,” Applied Sciences, vol. 14, no. 17, pp. 7447–7447, 2024, https://doi.org/10.3390/app14177447.
- L. Alzubaidi, M. A. Fadhel, F. Hollman, A. Salhi, J. Santamaria, Y. Duan, A. Gupta, K. Cutbush, A. Abbosh, and Y. Gu, “SSP: self-supervised pertaining technique for classification of shoulder implants in x-ray medical images: a broad experimental study,” Artificial Intelligence Review, vol. 57, no. 10, 2024, https://doi.org/10.1007/s10462-024-10878-0.
- C. Hong, M. Liu, D. M. Wojdyla, J. Hickey, M. Pencina, and R. Henao, “Trans-Balance: Reducing demographic disparity for prediction models in the presence of class imbalance,” Journal of biomedical informatics, vol. 149, p. 104532, 2024, https://doi.org/10.1016/j.jbi.2023.104532.
- M. N. Alnuaimi et la., “Transfer Learning Empowered Skin Diseases Detection in Children,” Computer Modeling in Engineering & Sciences, vol. 0, no. 0, pp. 1–10, 2024, https://doi.org/10.32604/cmes.2024.055303.
- P. Xiang, L. Zhou, and L. Tang, “Transfer learning via random forests: A one-shot federated approach,” Computational Statistics & Data Analysis, vol. 197, p. 107975, 2024, https://doi.org/10.1016/j.csda.2024.107975.
- O. Khouadja, M. S. Naceur, S. Mhamedi, and A. Baffoun, “Tunisian Lung Cancer Dataset: Collection, Annotation and Validation with Transfer Learning,” International Journal of Advanced Computer Science and Applications, vol. 15, no. 7, 2024, https://doi.org/10.14569/ijacsa.2024.01507121.
- A. S. Sambyal, U. Niyaz, N. C. Krishnan, and D. R. Bathula, “Understanding calibration of deep neural networks for medical image classification,” Computer Methods and Programs in Biomedicine, vol. 242, p. 107816, 2023, https://doi.org/10.1016/j.cmpb.2023.107816.
- S. Benbakreti, S. Benbakreti, K. Benyahia, and M. Benouis, “Using Resnet18 in A Deep-Learning Framework and Assessing the Effects of Adaptive Learning Rates in the Identification of Malignant Breast Masses in Mammograms,” Jordanian Journal of Computers and Information Technology, no. 0, pp. 1–1, 2024, https://doi.org/10.5455/jjcit.71-1699818406.
- S. K. Mathivanan, S. Sonaimuthu, S. Murugesan, H. Rajadurai, B. D. Shivahare, and M. A. Shah, “Employing deep learning and transfer learning for accurate brain tumor detection,” Scientific reports, vol. 14, no. 1, 2024, https://doi.org/10.1038/s41598-024-57970-7.
- Z. Alammar, L. Alzubaidi, J. Zhang, Y. Li, W. Lafta, and Y. Gu, “Deep Transfer Learning with Enhanced Feature Fusion for Detection of Abnormalities in X-ray Images,” Cancers, vol. 15, no. 15, pp. 4007–4007, 2023, https://doi.org/10.3390/cancers15154007.
- R. Salakapuri, P. V. Terlapu, K. R. Kalidindi, R. N. Balaka, D. Jayaram, and T. Ravikumar, “Intelligent brain tumor detection using hybrid finetuned deep transfer features and ensemble machine learning algorithms,” Scientific Reports, vol. 15, no. 1, 2025, https://doi.org/10.1038/s41598-025-08689-6.
- P. K. Mall, P. K. Singh, S. Srivastav, V. Narayan, M. Paprzycki, T. Jaworska, and M. Ganzha, “A comprehensive review of deep neural networks for medical image processing: Recent developments and future opportunities,” Healthcare Analytics, vol. 4, p. 100216, 2023, https://doi.org/10.1016/j.health.2023.100216.
- H. Sadr, et al., “Unveiling the potential of artificial intelligence in revolutionizing disease diagnosis and prediction: a comprehensive review of machine learning and deep learning approaches,” European journal of medical research, vol. 30, no. 1, 2025, https://doi.org/10.1186/s40001-025-02680-7.
- Y. Pamungkas and U. W. Astuti, "Comparison of Human Emotion Classification on Single-Channel and Multi-Channel EEG using Gate Recurrent Unit Algorithm," 2023 International Conference on Computer Science, Information Technology and Engineering (ICCoSITE), pp. 1-6, 2023, https://doi.org/10.1109/iccosite57641.2023.10127686.
- Y. Bian, J. Li, C. Ye, X. Jia, and Q. Yang, “Artificial intelligence in medical imaging: From task-specific models to large-scale foundation models,” Chinese Medical Journal, vol. 138, no. 06, pp. 651-663 2025, https://doi.org/10.1097/cm9.0000000000003489.
- J. G. d. Almeida, L. C. Alberich, G. Tsakou, K. Marias, M. Tsiknakis, K. Lekadir, L. M.-Bonmati, and N. Papanikolaou, “Foundation models for radiology—the position of the AI for Health Imaging (AI4HI) network,” Insights into Imaging, vol. 16, no. 1, 2025, https://doi.org/10.1186/s13244-025-02056-9.
- S. E. Abhadiomhen, E. O. Nzeakor, and K. Oyibo, “Health Risk Assessment Using Machine Learning: Systematic Review,” Electronics, vol. 13, no. 22, p. 4405, 2024, https://doi.org/10.3390/electronics13224405.
- J. Han, H. Zhang, and K. Ning, “Techniques for learning and transferring knowledge for microbiome-based classification and prediction: review and assessment,” Briefings in Bioinformatics, vol. 26, no. 1, 2024, https://doi.org/10.1093/bib/bbaf015.
- D. M. Anisuzzaman, J. G. Malins, P. A. Friedman, and Z. I. Attia, “Fine-Tuning LLMs for Specialized Use Cases,” Mayo Clinic Proceedings: Digital Health, vol. 3, no. 1, 2024, https://doi.org/10.1016/j.mcpdig.2024.11.005.
Yuri Pamungkas (Transfer Learning Models for Precision Medicine: A Review of Current Applications)