ISSN: 2685-9572 Buletin Ilmiah Sarjana Teknik Elektro
Vol. 7, No. 4, December 2025, pp. 1045-1056
An Innovation Approach for Feature Selection Medical Data Using Joint Fine-Tuning Fusion Graph Convolutional Network
Dimas Chaerul Ekty Saputra 1,2, Irianna Futri 3, Elvaro Islami Muryadi 4,5
1 Informatics Study Program, Telkom University, Surabaya Campus, Surabaya 60231, East Java, Indonesia
2 Center of Excellence for Motion Technology for Safety Health and Wellness, Research Institute of Sustainable Society, Telkom University, Surabaya Campus, Surabaya 60231, East Java, Indonesia
3 Department of International Technology and Innovation Management, International College, Khon Kaen University, Khon Kaen 40002, Thailand
4 Department of Community, Occupational, and Family Medicine, Faculty of Medicine, Khon Kaen University, Khon Kaen 40002, Thailand
5 Department of Public Health, Faculty of Health Sciences, Adiwangsa Jambi University, Jambi 36138, Indonesia
ARTICLE INFORMATION |
| ABSTRACT |
Article History: Received 29 August 2024 Revised 01 October 2024 Accepted 03 January 2026 |
| 
This research addresses the challenge of feature selection in high dimensional medical datasets, where unnecessary or duplicated information can hide patterns and negatively impact model performance. The aim is to develop an efficient feature selection strategy using Fine-tuning Fusion Graph Convolutional Networks (GCNs) to enhance model accuracy and interpretability. The objectives include improving the medical data selection process, increasing generalization, and assisting healthcare professionals in making educated clinical decisions based on the most relevant factors. The study employs Joint Fine-Tuning Fusion Graph Convolutional Networks (GCNs) for feature selection in medical datasets. This approach entails creating several graphs to illustrate feature interrelations, amalgamating them into a cohesive representation, and optimizing the model to emphasize pertinent aspects. The L2-norm of the final embeddings dictates feature significance, directing the choice of the most critical features for enhanced predictive accuracy. The study's findings indicate that GCN-based feature selection improves classification accuracy, especially for the PIDD dataset, enhancing accuracy, precision, recall, and F1-score from 0.74 to 0.75. The Kidney Failure dataset exhibited near-perfect accuracy (0.99) prior to selection, whereas the heart disease dataset had a minor reduction in performance (from 0.81 to 0.80), highlighting the dataset-specific effects of feature selection. GCN-based feature selection improved classification performance, increasing the PIDD dataset's accuracy from 0.74 to 0.75, with no significant effect on the Kidney Failure dataset. Nonetheless, it somewhat diminished performance for the heart disease dataset. Subsequent study ought to enhance feature selection techniques by integrating dataset-specific optimizations and domain expertise to augment model precision and overall generalizability. |
Keywords: Feature Selection; Graph Convolutional Networks; Medical Data; Joint Fine-Tuning; Clinical Decision Support |
Corresponding Author: Dimas Chaerul Ekty Saputra, Informatics Study Program, Telkom University, Surabaya Campus, Surabaya 60231, East Java, Indonesia Email:dimaschaerulekty@telkomuniersity.ac.id |
This work is open access under a Creative Commons Attribution-Share Alike 4.0 
|
Document Citation: D. C. E. Saputra, I. Futri, and E. I. Muryadi, “An Innovation Approach for Feature Selection Medical Data Using Joint Fine-Tuning Fusion Graph Convolutional Network,” Buletin Ilmiah Sarjana Teknik Elektro, vol. 7, no. 4, pp. 1045-1056, 2025, DOI: 10.12928/biste.v7i4.11652. |
- INTRODUCTION
The rapidly changing realm of healthcare data analysis requires creative methods to tackle the intrinsic complexity and extensive quantities of medical information [1]-[4]. With the emergence of electronic health records, wearable health devices, and sophisticated imaging techniques, healthcare organizations produce an abundance of data every day [5]-[9]. This surge of information has significant promises for improving patient care, optimizing clinical processes, and guiding evidence-based medical choices [10]-[14]. The issue resides not only in the substantial amount of data but also in its complexity, including varied formats, inconsistent data quality, and deep interrelationships across elements [15]-[18]. Consequently, there is an urgent need for new strategies capable of efficiently extracting relevant insights from this data, especially via complex feature selection procedures [19]-[22].
Feature selection is an essential phase in the data pretreatment pipeline, particularly for medical datasets marked by high dimensionality [23]-[26]. Conventional machine learning methods sometimes encounter difficulties with datasets that include an excessive amount of characteristics, many of which may be extraneous or superfluous [5],[15],[27][28]. The existence of such characteristics may mask significant patterns, complicate model training, and ultimately result in inferior prediction performance. Furthermore, the likelihood of overfitting escalates with a growth in the number of features, leading to models that do not generalize well to novel, unexplored data. This highlights the need to identify and choose the most relevant elements that substantially influence the goal results [20],[29]-[31].
This paper provides a unique method using Fine-tuning Fusion Graph Convolutional Networks (GCNs) for feature selection in medical data to address these problems [32]. This technique utilizes the distinctive characteristics of GCNs to improve the feature selection process, ensuring that only the most relevant features are preserved for model training. Graph Convolutional Networks (GCNs) are especially adept at modeling relationships among elements within a graph framework [33]-[36]. In the realm of medical data, this entails depicting characteristics and patient interactions as nodes and edges in a graph, enabling the model to include both the individual importance of each feature and the contextual meaning derived from their interrelations [37][38].
This study is primarily motivated by the increasing need for precise and interpretable prediction models in healthcare [39]. As healthcare practitioners increasingly depend on data-driven decision-making, the need for openness in model projections becomes essential [40]. Healthcare practitioners must comprehend and rely on the elements affecting forecasts to make educated clinical judgments [41]. Conventional black-box models, while theoretically precise, sometimes lack the requisite interpretability for clinical use [42]. This study seeks to improve model interpretability by including feature selection into the GCN framework, enabling practitioners to discern the most significant factors influencing the model's predictions. This congruence with clinical requirements enhances the model's applicability in actual environments and promotes increased acceptability of machine learning methods among healthcare practitioners.
The objectives of this study are diverse. Initially, it aims to provide an efficient feature selection process specifically designed for the distinct attributes of high-dimensional medical datasets. This entails the design and construction of a Fine-tuning Fusion GCN that may adaptively modify to the distinct characteristics of the data, enhancing the selection process according to the context of the medical application. The study seeks to improve the interpretability of model predictions, offering healthcare professionals clear insights into the most significant elements for clinical decision-making. This work aims to illustrate the practical applicability of the suggested strategy via extensive experiments using real-world medical information, highlighting its potential to enhance patient outcomes and facilitate evidence-based practices.
This study is distinguished by its use of Fine-tuning Fusion approaches inside the GCN framework, differentiating it from conventional feature selection methods. Although much research has investigated GCNs for many applications, the concentrated emphasis on feature selection in medical data signifies a notable progression in the domain. The suggested technique utilizes a fine-tuning process to adjust the model to various medical datasets, enhancing the comprehension of feature significance according to the distinct context of each dataset. This adaptive modification improves the model's capacity to comprehend the intricacies of medical data, yielding more precise and relevant feature choices.
Furthermore, the suggested Fine-tuning Fusion GCN methodology tackles the computational efficiency of handling extensive medical data, a vital factor in the contemporary healthcare landscape [43]. As healthcare organizations progressively embrace data-driven approaches, the need for real-time analysis and decision-making is increasing [44]. This study seeks to integrate the advantages of GCNs with fine-tuning methodologies to provide a system that harmonizes computing efficiency and accuracy in feature selection. This capacity is especially significant in clinical environments as prompt insights may directly influence patient care, allowing healthcare practitioners to make educated choices based on the most relevant data.
This research presents significant advancements in medical data analysis, including:
- A novel methodology that integrates Fine-tuning and Fusion techniques within Graph Convolutional Networks (GCNs) to improve feature selection, establishing a robust framework for medical data analysis.
- Enhanced model performance by efficient feature selection, minimizing overfitting, and improving generalization to novel data.
- Enhanced interpretability of model results, promoting improved clinical decision-making by pinpointing the most important elements for healthcare professionals.
- Empirical insights derived from actual medical data applications illustrate the efficacy of the suggested methodology in enhancing patient outcomes.
This study is motivated by the urgent need for efficient feature selection techniques specifically designed for medical data to address the issues of high dimensionality and inter-feature correlations. This research proposes an innovative technique that increases feature selection procedures by using Fine-tuning Fusion Graph Convolutional Networks, resulting in enhanced model performance and more informed clinical judgments. The next parts of this study will outline the utilized methodology, the performed experiments, and the acquired results, concluding with a discussion on the relevance of these findings for future research on the subject.
- LITERATURE REVIEW
The rapid development of artificial intelligence (AI), graph-based learning, and multimodal data integration has significantly advanced computational methods in healthcare and medical informatics [45]-[48]. Recent research demonstrates that graph-based models are particularly effective for capturing relational structures that traditional statistical or deep learning approaches often overlook. These models support the representation of complex interactions, whether among herbs in Traditional Chinese Medicine (TCM), salient features in imaging tasks, or multimodal patient information. As a result, graph-based learning increasingly contributes to improvements in predictive accuracy and clinical interpretability.
In the context of TCM, herbal prescription formulation is grounded in centuries of accumulated knowledge. However, many computational models used in earlier studies relied on simplified statistical representations such as bag-of-words, which fail to capture the nuanced relationships between herbs and symptoms. To address this issue, Yang et al. introduced a graph convolutional network (GCN) equipped with multi-layer information fusion that incorporates herb knowledge graphs into the learning process [49]. Their approach enriches feature representation, reduces noise, and improves predictive performance. The model achieved higher Precision (an increase of 6.2 percent), Recall (an increase of 16.0 percent), and F1-score (an increase of 12.0 percent) compared with baseline methods. These results highlight the growing potential of AI-driven tools to support prescription analysis and clinical decision-making in TCM.
Graph-based learning has also contributed to advancements in computer vision tasks such as co-saliency detection, where the objective is to identify shared salient regions across multiple images. Traditional methods often struggle to capture higher-level semantic consistencies. Hu et al. addressed this limitation by proposing a multi-scale graph fusion framework that integrates VGG-16-based feature extraction with GCN-based refinement [34]. Their method segments images into semantic superpixels and aggregates shared and scale-specific information to enhance discriminative capability. Evaluations on three benchmark datasets show that the model consistently outperforms leading approaches in all major metrics, demonstrating improved robustness and accuracy.
Graph Convolutional Networks have further proven useful in clinical prediction models, particularly in domains where datasets exhibit substantial variability. In coronary heart disease (CHD) prediction, existing GNN-based models often lack generalizability across datasets due to domain-specific characteristics. Lin et al. introduced a domain-adaptive multichannel GCN (DAMGCN) to overcome these challenges [50]. The framework combines dual-channel graph convolution, attention-guided node representation, and domain adversarial learning to align the feature distributions of source and target datasets. Through the joint optimization of several loss functions, the model successfully transfers knowledge between CHD datasets with differing properties. Experimental results confirm that DAMGCN performs significantly better than previous models, with additional improvements observed when graph-based transfer learning is incorporated.
Multimodal disease prediction represents another important application of graph-based reasoning. Traditional convolutional neural networks (CNNs) excel at processing imaging data but face challenges when integrating heterogeneous data sources. To address this, Huang and Chung proposed an edge-variational GCN that constructs dynamic population graphs and optimizes their structure simultaneously with spectral GCN layers [51]. The model also employs Monte-Carlo edge dropouts to enhance uncertainty estimation. Evaluations across four multimodal datasets related to conditions such as autism spectrum disorder and Alzheimer’s disease demonstrate notable accuracy improvements over earlier methods. Comprehensive ablation analysis further validates the importance of each architectural component.
In oncology, accurate survival prediction relies heavily on multimodal information that includes clinical, pathological, and genetic data. However, missing modalities are common and can severely reduce model reliability. Hou et al. developed a Hybrid Graph Convolutional Network (HGCN) to address this challenge [52]. The framework integrates GCNs with hypergraph convolutional networks (HCNs) and incorporates an online masked autoencoder to reconstruct missing hyperedges. This approach enables more effective modeling of intra- and inter-modal relationships. Experiments conducted on six cancer cohorts from The Cancer Genome Atlas (TCGA) reveal that HGCN significantly surpasses state-of-the-art methods, both in complete and incomplete modality scenarios, underscoring its practical utility in precision oncology.
Overall, the reviewed studies illustrate how graph-based learning methods continue to transform medical data analysis. From herb recommendation systems in TCM to visual saliency detection, cross-domain clinical prediction, multimodal disease modeling, and cancer survival analysis, recent research highlights the capacity of graph-centered approaches to generate more accurate, robust, and clinically meaningful insights.
- JOINT FINE-TUNING FUSION GRAPH CONVOLUTIONAL NETWORK
The joint fine-tuning fusion of Graph Convolutional Networks (GCNs) generally denotes a procedure in which several components of the model, or numerous models, are concurrently refined to enhance performance. This may include the concurrent optimization of several components of a GCN for tasks such as node classification, graph classification, or feature selection. The objective is to enable the GCN to acquire an improved representation by concurrently optimizing the architecture and characteristics of various components of the network, often while incorporating numerous sources of information or graphs. The Joint Fine-Tuning Fusion Graph Convolutional Network (GCN) for feature selection seeks to integrate information from several graphs that depict distinct connections among features while optimizing the model via fine-tuning fusion. This method integrates these connections inside the GCN framework to prioritize characteristics according to their significance in forecasting the target variable
The process begins with the construction of multiple graphs 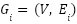 , where V represents the feature nodes and
, where V represents the feature nodes and 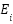 represents edges that encode relationships between the features. Each graph corresponds to a different type of feature relationship. For instance, a correlation graph would have edges weighted by the Pearson correlation coefficient,
represents edges that encode relationships between the features. Each graph corresponds to a different type of feature relationship. For instance, a correlation graph would have edges weighted by the Pearson correlation coefficient, 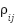 , between two features
, between two features 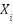 and
and 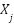 . Similarly, a mutual information graph would have edges weighted by the mutual information
. Similarly, a mutual information graph would have edges weighted by the mutual information 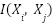 between features.
between features.
The adjacency matrix for each graph 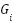 is represented by
is represented by 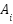 , and the feature matrix
, and the feature matrix 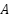 represents the feature values across all samples, where
represents the feature values across all samples, where 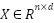 , with 𝑛 being the number of samples and 𝑑 the number of features.
, with 𝑛 being the number of samples and 𝑑 the number of features.
In the GCN, each layer propagates information between connected nodes (features) in a graph. This process is described by the following equation:
| 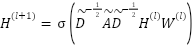
| (1) |
where 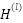 is the feature representation at layer 𝑙,
is the feature representation at layer 𝑙, 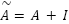 is the adhacency matrix with added self-loops to include each feature’s own information,
is the adhacency matrix with added self-loops to include each feature’s own information, 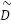 is the degree matrix derived from
is the degree matrix derived from 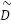 ,
, 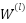 is the weight matrix at layer 𝑙, and
is the weight matrix at layer 𝑙, and 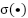 is an activation function.
is an activation function.
This formula aggregates information from a feature's neighbors in the graph at each layer, allowing the model to learn more complex relationships.
To integrate information from multiple graphs, a fusion mechanism is employed. The node representations (features) obtained from each graph 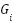 after applying (1) are fused into a unified representation. The fusion is typically a weighted sum of the individual graph representations:
after applying (1) are fused into a unified representation. The fusion is typically a weighted sum of the individual graph representations:
| 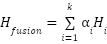
| (2) |
where 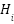 is the node representation from the
is the node representation from the 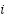 -th graph,
-th graph, 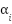 is a learnable weight parameter representing the importance of the
is a learnable weight parameter representing the importance of the 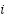 -th graph, and 𝑘 is the number of graphs.
-th graph, and 𝑘 is the number of graphs.
The weighted sum ensures that the most informative graphs contribute more to the final feature representation.
After constructing the fused representation, the model undergoes fine-tuning. This process adjusts the weights 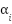 for each graph to prioritize the most important graph structures. The loss function for joint fine- tuning consists of two terms:
for each graph to prioritize the most important graph structures. The loss function for joint fine- tuning consists of two terms:
| 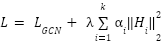
| (3) |
where 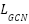 is the GCN loss,
is the GCN loss, 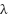 is the regularization parameter, and the second term is a regularization on the graph fusion weights
is the regularization parameter, and the second term is a regularization on the graph fusion weights 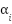 , which prevents overfitting by controlling the influence of each graph.
, which prevents overfitting by controlling the influence of each graph.
This joint optimization process ensures that the GCN not only learns to predict the target variable but also identifies the most important graph structures for feature selection.
Once the GCN is fine-tuned, feature importance is calculated based on the final node embeddings 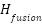 . The importance of each feature is determined by the L2-norm of its embedding in the final fused graph representation:
. The importance of each feature is determined by the L2-norm of its embedding in the final fused graph representation:
| 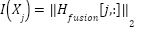
| (4) |
This score measures the influence of each feature in the context of all the graph structures. Features with higher importance scores are considered more relevant for predicting the target variable.
Utilizing the feature importance scores from (4), the top 𝑚 features may be identified by ranking them in decreasing order of significance. This last selection phase guarantees that only the most relevant attributes are used for subsequent classification assignments.
The Joint Fine-Tuning Fusion Graph Convolutional Network for feature selection integrates the advantages of numerous graphs, each representing distinct connections among features. The GCN consolidates data from these graphs (as defined in (1)), integrates the graph representations (as shown in (2)), and refines the model to enhance the feature ranking procedure (as described in (3)). The L2-norm of the final node embeddings (obtained from (4)) is used to compute feature significance, facilitating a reliable and precise identification of the most significant features.
- EXPERIMENTAL RESULTS AND DISCUSSIONS
All datasets and algorithms in this study were conducted on a single laptop, specifically the MacBook Pro 2020 M1, utilizing the Python computational platform to ensure equitable comparisons. This work uses feature selection techniques to enhance classification accuracy by obtaining kernel feature subsets. The Support Vector Machine (SVM) is utilized as the classification method to assess the subsets derived from feature selection. The quantity of features chosen for classification is contingent upon the classification's accuracy. Numerous established or extensively utilized feature selection approaches are available for comparison. Table 1 details the data that we used in this research.
Table 1. Data Information
Dataset | # Features | # Classes | # Instances |
PIDD | 9 | 2 | 768 |
Kidney Failure | 26 | 2 | 400 |
Heart Disease | 14 | 2 | 1,025 |
Figure 1 illustrates a consolidated graph of attributes from the Pima Indians Diabetes Dataset (PIDD), with each node representing a characteristic and the edges denoting the links or connections among these features. This visualization is crucial to the feature selection process of a Graph Convolutional Network (GCN). In this context, the GCN examines the graph structure to discern and assess the significance of features based on their interconnections. Essential attributes like BMI, Glucose, Blood Pressure, Insulin, and Diabetes Pedigree Function are pivotal in the graph, demonstrating their significant correlations with other variables such as Skin Thickness, Age, and Outcome. GCN utilizes these linkages to assess the significance of each feature for categorization objectives. By analyzing the interrelatedness of features, the GCN can ascertain which features should be preserved and which may be superfluous or less informative.
In feature selection, the GCN identifies patterns in feature connectivity. If specific variables exhibit high centrality or are interconnected with several others, they are likely to convey significant information for predicting the result (diabetes). Conversely, traits with limited or weaker connections may be deemed less pertinent and could be recommended for elimination. The model of GCN optimizes the selection of informative features, diminishes dimensionality, and enhances the performance of the SVM model, as demonstrated by the marginal improvement in measures like accuracy, precision, recall, and F1-score following feature selection for the PIDD dataset.
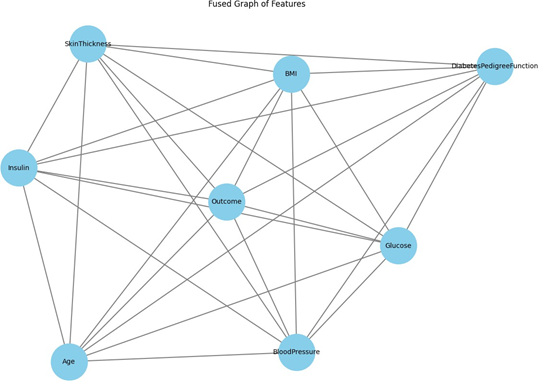
Figure 1. Graph Visualization from Diabetes Dataset (PIDD)
Figure 2 presents a consolidated graph of features for a kidney failure dataset, where feature selection utilizing a Graph Convolutional Network (GCN) entails the identification and retention of the most pertinent characteristics that substantially influence the classification process. Every node in the graph signifies a distinct feature pertinent to kidney health, including age, blood pressure, and blood urea, among others, and the edges connecting the nodes denote interactions or correlations among these features. The core node designated "classification" signifies the goal variable, indicating the presence or absence of renal failure. The GCN analyzes the graph by doing convolution operations on the nodes, wherein each node consolidates information from its adjacent nodes. This recurrent procedure enables the GCN to refine feature representations, thereby capturing significant patterns and dependencies among the features. The GCN discerns the importance of each feature concerning the classification task through various layers of convolution and aggregation. Features that demonstrate stronger associations or proximity to the primary classification node are often regarded as more significant. The concluding phase of feature selection entails preserving features with elevated learned importance scores, essential for precise classification while eliminating less significant features to diminish model complexity and improve interpretability. Key factors found for predicting kidney failure may encompass blood pressure, blood urea, serum creatinine, age, diabetes mellitus, and hemoglobin levels. The GCN accentuates these features because of their significant impact on the target variable, therefore enhancing the overall efficacy and dependability of the classification model.
Figure 3 illustrates a consolidated graph of attributes for the heart disease dataset. This graph depicts nodes representing features (e.g., cp, thal, trestbps, target), with edges indicating the interconnections among these features, demonstrating their interactions. This depiction is integral to the feature selection process utilizing GCN, which aids in identifying the most significant features for predicting heart disease. Crucial attributes such as cp (chest pain), thal (thalassemia), trestbps (resting blood pressure), and ca (number of main vessels) exhibit a substantial level of interconnection, signifying their importance in influencing the outcome or target variable. The center node target presumably signifies the classification result (presence or absence of heart disease), and its numerous connections indicate that several features directly influence the prediction process.
In the realm of feature selection, the GCN examines these interactions to determine the most essential features for model correctness and those that are superfluous. Attributes having robust and many interconnections to others, such as thalach (highest heart rate achieved), chol (cholesterol level), and oldpeak (ST depression induced by exercise), may possess considerable predictive capability. Conversely, attributes having limited or weaker associations, such as sex and fbs (fasting blood sugar), may be regarded as less essential for the categorization task. Employing GCNs for feature selection enables the reduction of dataset dimensionality without compromising model performance, facilitating a more efficient and precise categorization of heart disease. This technique improves the model's generalizability by emphasizing the most useful traits and eliminating those that contribute minimally to the overall prediction.
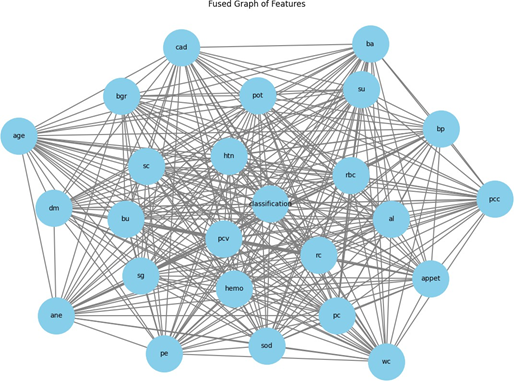
Figure 2. Graph Visualization from Kidney Failure Dataset
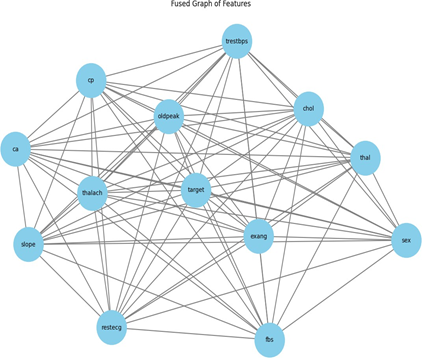
Figure 3. Graph Visualization from Heart Disease
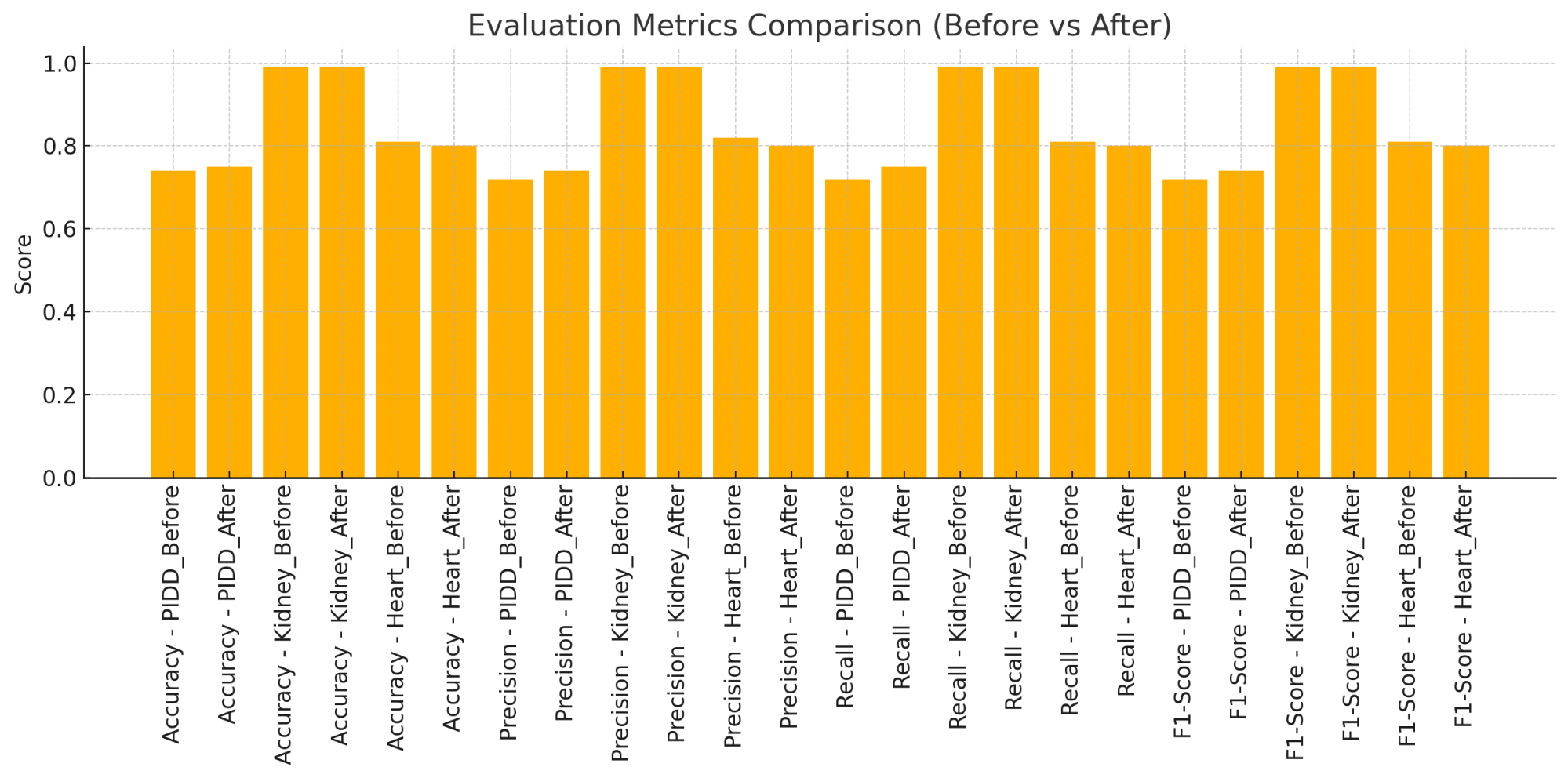
The assessment of the SVM classifier's efficacy before and after feature selection via a Graph Convolutional Network (GCN) across three datasets—PIDD, Kidney Failure, and Heart Disease—yields significant insights into the influence of feature selection on model performance shows in Table 2 and illustrated in Figure 4. Utilizing GCN-based feature selection for the PIDD (Pima Indians Diabetes Dataset) resulted in a significant enhancement in all assessment measures. The accuracy rose from 0.74 to 0.75, while precision, recall, and F1-score improved from 0.72 to 0.74 and 0.75, respectively. This indicates that feature selection successfully diminished noise and eliminated extraneous features, enabling the model to concentrate on more pertinent features. Consequently, the SVM classifier's capacity to differentiate between classes improved marginally, suggesting that the feature selection method augmented the overall quality of the input data.
In the Kidney Failure dataset, the SVM classifier exhibited consistently good performance, achieving accuracy, precision, recall, and F1-score of 0.99 both before and during feature selection. This outcome signifies that the initial feature collection was already significantly pertinent and informative for this specific classification job, exhibiting minimal noise or extraneous features. The feature selection approach did not significantly impact the model's performance, and the classifier achieved nearly flawless results. This indicates that in specific datasets with clearly defined features, feature selection may not produce substantial enhancements, yet it also does not impair performance.
Table 2. Evaluation Metrics from each Dataset
Dataset | Comparison | Evaluation Metrics |
Accuracy | Precision | Recall | F1-Score |
PIDD | Before | 0.74 | 0.72 | 0.72 | 0.72 |
After | 0.75 | 0.74 | 0.75 | 0.74 |
Kidney Failure | Before | 0.99 | 0.99 | 0.99 | 0.99 |
After | 0.99 | 0.99 | 0.99 | 0.99 |
Heart Disease | Before | 0.81 | 0.82 | 0.81 | 0.81 |
After | 0.8 | 0.8 | 0.8 | 0.8 |

Figure 4. Visualization from Evaluation Metrics for each Dataset
The performance of the SVM classifier somewhat diminished following feature selection in the heart disease dataset. The accuracy declined from 0.81 to 0.80, accompanied by reductions in precision, recall, and F1-score from 0.81 and 0.82 to 0.80. The minor decline in performance indicates that the GCN-based feature selection may have omitted certain pertinent features crucial for the classifier. This result underscores the inherent risk in feature selection techniques: excessive removal of features or the exclusion of essential elements may result in diminished model performance. Consequently, it emphasizes the necessity of meticulous selection and calibration of feature selection techniques to guarantee the elimination of only genuinely redundant or uninformative characteristics. GCN-based feature selection exhibits varied effects contingent upon the dataset. In datasets such as PIDD, feature selection enhances model performance by minimizing noise and emphasizing more informative characteristics. Nonetheless, for datasets with clearly delineated feature sets, such as Kidney Failure, feature selection may have minimal to no impact. Conversely, in instances such as the heart disease dataset, improper feature selection may result in a marginal decline in performance, underscoring the necessity for dataset-specific optimization of the feature selection process. This analysis highlights the context-dependent characteristics of feature selection and the necessity for comprehensive examination before its use across various datasets.
- CONCLUSIONS
This research illustrates the differential efficacy of GCN-based feature selection across several medical datasets. In the Pima Indians Diabetes Dataset (PIDD), the application of GCN for feature selection markedly enhanced the efficacy of the Support Vector Machine (SVM) classifier. The accuracy rose from 0.74 to 0.75, while precision, recall, and F1-score enhanced from 0.72 to 0.74 and 0.75, respectively. This enhancement demonstrates that GCN-based feature selection efficiently diminished noise and eliminated extraneous features, enabling the model to concentrate on more pertinent variables such as BMI, Glucose, Blood Pressure, and Insulin. This indicates that GCN is especially advantageous for datasets characterized by significant interconnection among characteristics, hence enhancing classification accuracy by prioritizing the most essential variables. Conversely, the feature selection method had minimal to no effect on the Kidney Failure dataset. The SVM classifier attained nearly flawless results both prior to and after to feature selection, sustaining accuracy, precision, recall, and F1-score of 0.99. This suggests that the initial feature set was already highly appropriate for classification, with negligible noise or redundant features. Consequently, the feature selection did not markedly improve performance, indicating that for datasets with a well-defined and pertinent feature space, further feature selection may be superfluous.
Nonetheless, in the instance of the heart disease dataset, a marginal decline in performance was noted following feature selection. The accuracy decreased from 0.81 to 0.80, accompanied by minor declines in precision, recall, and F1-score. This reduction indicates that the GCN-based selection procedure may have unintentionally eliminated vital features necessary for the classification task, resulting in diminished model performance. This result highlights the possible dangers of excessive filtering and the necessity for meticulous optimization and validation of feature selection methods to prevent the omission of relevant features. Future research should investigate more sophisticated fine-tuning methodologies for GCN-based feature selection, emphasizing the equilibrium between eliminating redundant characteristics and preserving useful ones. Furthermore, the creation of hybrid models that integrate GCN-based feature selection with alternative machine learning methodologies, such as ensemble techniques, may yield a more resilient framework for managing varied datasets. Future study should explore the efficacy of GCN-based feature selection on larger and more intricate datasets, possibly including multi-class classification tasks or time-series data, to enhance understanding of its generalizability. Furthermore, investigating the incorporation of domain expertise, especially within medical datasets, may facilitate the feature selection process, guaranteeing the retention of critical features informed by expert knowledge. Future research should focus on optimizing and adapting GCN- based feature selection for diverse applications, enhancing classification accuracy and model robustness across a broader spectrum of datasets.
Acknowledgement
This research was partially supported by the Informatics Study Program, Telkom University, Surabaya Campus, Indonesia. The authors also acknowledge the valuable assistance provided by the Center of Excellence for Motion Technology for Safety Health and Wellness, Research Institute of Sustainable Society, Telkom University, Surabaya Campus, Indonesia.
Conflicts of Interest
The authors declare no conflict of interest.
REFERENCES
- C. M. Cutillo et al., “Machine intelligence in healthcare—perspectives on trustworthiness, explainability, usability, and transparency,” Npj Digit. Med., vol. 3, no. 1, p. 47, 2020, https://doi.org/10.1038/s41746-020-0254-2.
- E. Y. Boateng and D. A. Abaye, “A Review of the Logistic Regression Model with Emphasis on Medical Research,” J. Data Anal. Inf. Process., vol. 07, no. 04, pp. 190–207, 2019, https://doi.org/10.4236/jdaip.2019.74012.
- A. Alzubaidi, “Challenges in Developing Prediction Models for Multi-modal High-Throughput Biomedical Data,” in Intelligent Systems and Applications, vol. 868, 1056–1069, 2019, https://doi.org/10.1007/978-3-030-01054-6_73.
- B. Çil, H. Ayyıldız, and T. Tuncer, “Discrimination of β-thalassemia and iron deficiency anemia through extreme learning machine and regularized extreme learning machine based decision support system,” Med. Hypotheses, vol. 138, p. 109611, 2020, https://doi.org/10.1016/j.mehy.2020.109611.
- [5] D. C. E. Saputra, K. Sunat, and T. Ratnaningsih, “A new artificial intelligence approach using extreme learning machine as the potentially effective model to predict and analyze the diagnosis of anemia,” Healthcare, vol. 11, no. 5, p. 697, 2023, https://doi.org/10.3390/healthcare11050697.
- D. C. E. Saputra, Y. Maulana, E. Faristasari, A. Ma’arif, and I. Suwarno, “Machine Learning Performance Analysis for Classification of Medical Specialties,” in Proceeding of the 3rd International Conference on Electronics, Biomedical Engineering, and Health Informatics, vol. 1008, pp. 513–528, 2023, https://doi.org/10.1007/978-981-99-0248-4_34.
- C. Guo and J. Chen, “Big Data Analytics in Healthcare,” in Knowledge Technology and Systems, vol. 34, pp. 27–70, 2023, https://doi.org/10.1007/978-981-99-1075-5_2.
- M. M. Chowdhury, R. S. Ayon, and M. S. Hossain, “An investigation of machine learning algorithms and data augmentation techniques for diabetes diagnosis using class imbalanced BRFSS dataset,” Healthc. Anal., vol. 5, p. 100297, 2024, https://doi.org/10.1016/j.health.2023.100297.
- M. Ağraz, E. Eğrioğlu, E. Baş, M.-Y. Chen, D. Göksülük, and M. F. Burak, “Diabetes Development Prediction Using a Hybrid Model Combining Dendritic Artificial Neuron Model and Logistic Regression,” Endocrinol. Res. Pract., vol. 29, no. 2, pp. 84–93, 2025, https://doi.org/10.5152/erp.2025.24585.
- C. Prod’homme et al., “Can palliative care consultation increase integration of palliative care for patients with hematologic malignancies?,” Blood Adv., vol. 5, no. 8, pp. 2123–2127, 2021, https://doi.org/10.1182/bloodadvances.2021004369.
- P. Martens, P. Nijst, F. H. Verbrugge, K. Smeets, M. Dupont, and W. Mullens, “Impact of iron deficiency on exercise capacity and outcome in heart failure with reduced, mid-range and preserved ejection fraction,” Acta Cardiol., vol. 73, no. 2, pp. 115–123, 2018, https://doi.org/10.1080/00015385.2017.1351239.
- P. Y. Taser, “Application of Bagging and Boosting Approaches Using Decision Tree-Based Algorithms in Diabetes Risk Prediction,” in The 7th International Management Information Systems Conference, p. 6, 2021, https://doi.org/10.3390/proceedings2021074006.
- M. Kashina, I. D. Lenivtceva, and G. D. Kopanitsa, “Preprocessing of unstructured medical data: the impact of each preprocessing stage on classification,” Procedia Comput. Sci., vol. 178, pp. 284–290, 2020, https://doi.org/10.1016/j.procs.2020.11.030.
- D. C. E. Saputra, E. I. Muryadi, R. Phann, I. Futri, and L. Lismawati, “An Innovative Artificial Intelligence-Based Extreme Learning Machine Based on Random Forest Classifier for Diagnosed Diabetes Mellitus,” J. Ilm. Tek. Elektro Komput. Dan Inform., vol. 10, no. 1, pp. 173–187, 2024, https://doi.org/10.26555/jiteki.v10i1.28690.
- D. C. E. Saputra, Y. Maulana, T. A. Win, R. Phann, and W. Caesarendra, “Implementation of Machine Learning and Deep Learning Models Based on Structural MRI for Identification of Autism Spectrum Disorder,” vol. 9, no. 2, 2023, https://doi.org/10.26555/jiteki.v9i2.26094.
- Z. Cai, H. Huang, G. Sun, ZiQiang. Li, and ChengJu. Ouyang, “Advancing Predictive Models: Unveiling LightGBM Machine Learning for Data Analysis,” in 2023 4th International Conference on Computer, Big Data and Artificial Intelligence (ICCBD+AI), pp. 109–112, 2023, https://doi.org/10.1109/iccbd-ai62252.2023.00027.
- V. R. S. Kushwah and K. Verma, “Security and Privacy Challenges for Big Data on Social Media,” in Big Data Analytics in Cognitive Social Media and Literary Texts, pp. 267–285, 2021, https://doi.org/10.1007/978-981-16-4729-1_15.
- U. Jain, S. Kumar, S. Dubey, O. Sharma, and V. Kumar Jain, “Virtualization-A New Dimension of Big Data,” in 2018 8th International Conference on Cloud Computing, Data Science & Engineering (Confluence), pp. 803–808, 2018, https://doi.org/10.1109/CONFLUENCE.2018.8443075.
- D. Albashish, A. I. Hammouri, M. Braik, J. Atwan, and S. Sahran, “Binary biogeography-based optimization based SVM-RFE for feature selection,” Appl. Soft Comput., vol. 101, p. 107026, 2021, https://doi.org/10.1016/j.asoc.2020.107026.
- S. Sivaranjani, S. Ananya, J. Aravinth, and R. Karthika, “Diabetes Prediction using Machine Learning Algorithms with Feature Selection and Dimensionality Reduction,” in 2021 7th International Conference on Advanced Computing and Communication Systems (ICACCS), pp. 141–146, 2021, https://doi.org/10.1109/ICACCS51430.2021.9441935.
- Y. Aggarwal, J. Das, P. M. Mazumder, R. Kumar, and R. K. Sinha, “Heart rate variability features from nonlinear cardiac dynamics in identification of diabetes using artificial neural network and support vector machine,” Biocybern. Biomed. Eng., vol. 40, no. 3, pp. 1002–1009, 2020, https://doi.org/10.1016/j.bbe.2020.05.001.
- J. V. N. Ramesh, A. Kushwaha, T. Sharma, A. Aranganathan, A. Gupta, and S. K. Jain, “Intelligent Feature Engineering and Feature Selection Techniques for Machine Learning Evaluation,” in Mobile Radio Communications and 5G Networks, vol. 915, pp. 753–764, 2024, https://doi.org/10.1007/978-981-97-0700-3_56.
- M. Y. M. Parvees and M. Raja, “Optimal Feature Subset Selection with Multi-Kernel Extreme Learning Machine for Medical Data Classification,” Turk. J. Comput. Math. Educ., vol. 12, no. 6, pp. 3612–3623, 2021, https://doi.org/https://doi.org/10.17762/turcomat.v12i6.7157.
- S. Li, X. Huang, and D. Wang, “Stochastic configuration networks for multi-dimensional integral evaluation,” Inf. Sci., vol. 601, pp. 323–339, 2022, https://doi.org/10.1016/j.ins.2022.04.005.
- S. Asghari, H. Nematzadeh, E. Akbari, and H. Motameni, “Mutual information-based filter hybrid feature selection method for medical datasets using feature clustering,” Multimed. Tools Appl., vol. 82, no. 27, pp. 42617–42639, 2023, https://doi.org/10.1007/s11042-023-15143-0.
- I. Hussain, M. Qureshi, M. Ismail, H. Iftikhar, J. Zywiołek, and J. L. López-Gonzales, “Optimal features selection in the high dimensional data based on robust technique: Application to different health database,” Heliyon, vol. 10, no. 17, p. e37241, 2024, https://doi.org/10.1016/j.heliyon.2024.e37241.
- G. Sun, C. Jiang, X. Wang, and X. Yang, “Short-term building load forecast based on a data-mining feature selection and LSTM-RNN method,” IEEJ Trans. Electr. Electron. Eng., vol. 15, no. 7, pp. 1002–1010, 2020, https://doi.org/10.1002/tee.23144.
- T. A. Al-Qablan, M. H. Mohd Noor, M. A. Al-Betar, and A. T. Khader, “Improved gray wolf harris hawk algorithm based feature selection for sentiment analysis,” Results Control Optim., vol. 20, p. 100604, 2025, https://doi.org/10.1016/j.rico.2025.100604.
- A. Bilal, G. Sun, S. Mazhar, and A. Imran, “Improved Grey Wolf Optimization-Based Feature Selection and Classification Using CNN for Diabetic Retinopathy Detection,” in Evolutionary Computing and Mobile Sustainable Networks, vol. 116, pp. 1–14, 2022, https://doi.org/10.1007/978-981-16-9605-3_1.
- H. Liu, M. Zhou, and Q. Liu, “An embedded feature selection method for imbalanced data classification,” IEEECAA J. Autom. Sin., vol. 6, no. 3, pp. 703–715, 2019, https://doi.org/10.1109/JAS.2019.1911447.
- A. Helisa, T. H. Saragih, I. Budiman, F. Indriani, and D. Kartini, “Prediction of Post-Operative Survival Expectancy in Thoracic Lung Cancer Surgery Using Extreme Learning Machine and SMOTE,” J. Ilm. Tek. Elektro Komput. Dan Inform., vol. 9, no. 2, pp. 239–249, 2023, https://doi.org/10.26555/jiteki.v9i2.25973.
- H. Li, X. Shi, X. Zhu, S. Wang, and Z. Zhang, “FSNet: Dual Interpretable Graph Convolutional Network for Alzheimer’s Disease Analysis,” IEEE Trans. Emerg. Top. Comput. Intell., vol. 7, no. 1, pp. 15–25, 2023, https://doi.org/10.1109/TETCI.2022.3183679.
- Y. Liu, H. Zhou, M. Guan, F. Feng, and J. Duan, “Scalp EEG-Based Automatic Detection of Epileptiform Events via Graph Convolutional Network and Bi-Directional LSTM Co-Embedded Broad Learning System,” IEEE Signal Process. Lett., vol. 30, pp. 448–452, 2023, https://doi.org/10.1109/LSP.2023.3263433.
- R. Hu, Z. Deng, and X. Zhu, “Multi-scale Graph Fusion for Co-saliency Detection.” Proc. AAAI Conf. Artif. Intell., vol. 35, no. 9, pp. 7789–7796, May 2021, https://doi.org/10.1609/aaai.v35i9.16951.
- J. Chen, B. Li, and K. He, “Neighborhood convolutional graph neural network,” Knowl.-Based Syst., vol. 295, p. 111861, 2024, https://doi.org/10.1016/j.knosys.2024.111861.
- V. Purna Chandra Reddy and K. K. Gurrala, “OHGCNet: Optimal feature selection-based hybrid graph convolutional network model for joint DR-DME classification,” Biomed. Signal Process. Control, vol. 78, p. 103952, 2022, https://doi.org/10.1016/j.bspc.2022.103952.
- Z. Xu, D. Yu, H. Hou, W. Zhang, and Y. Zhang, “Research on Fault Diagnosis of Rolling Bearing in Printing Press Based on Convolutional Neural Network,” in Advances in Graphic Communication, Printing and Packaging Technology and Materials, vol. 754, pp. 487–494, 2021, https://doi.org/10.1007/978-981-16-0503-1_71.
- J. Zheng, Z. Gao, J. Ma, J. Shen, and K. Zhang, “Deep Graph Convolutional Networks for Accurate Automatic Road Network Selection,” ISPRS Int. J. Geo-Inf., vol. 10, no. 11, p. 768, 2021, https://doi.org/10.3390/ijgi10110768.
- H. Sadr et al., “Unveiling the potential of artificial intelligence in revolutionizing disease diagnosis and prediction: a comprehensive review of machine learning and deep learning approaches,” Eur. J. Med. Res., vol. 30, no. 1, p. 418, May 2025, https://doi.org/10.1186/s40001-025-02680-7.
- M. Javaid, A. Haleem, and R. P. Singh, “Health informatics to enhance the healthcare industry’s culture: An extensive analysis of its features, contributions, applications and limitations,” Inform. Health, vol. 1, no. 2, pp. 123–148, 2024, https://doi.org/10.1016/j.infoh.2024.05.001.
- J. M. Schwartz et al., “Factors Influencing Clinician Trust in Predictive Clinical Decision Support Systems for In-Hospital Deterioration: Qualitative Descriptive Study,” JMIR Hum. Factors, vol. 9, no. 2, p. e33960, 2022, https://doi.org/10.2196/33960.
- M. Ennab and H. Mcheick, “Enhancing interpretability and accuracy of AI models in healthcare: a comprehensive review on challenges and future directions,” Front. Robot. AI, vol. 11, p. 1444763, 2024, https://doi.org/10.3389/frobt.2024.1444763.
- M. I. Mazdadi, T. H. Saragih, I. Budiman, A. Farmadi, and A. Tajali, “The Effectiveness of Data Imputations on Myocardial Infarction Complication Classification Using Machine Learning Approach with Hyperparameter Tuning,” J. Ilm. Tek. Elektro Komput. Dan Inform., vol. 10, no. 3, pp. 520–533, 2024, https://doi.org/10.26555/jiteki.v10i3.29479.
- G. Lyu, “Data-driven decision making in patient management: a systematic review,” BMC Med. Inform. Decis. Mak., vol. 25, no. 1, p. 239, 2025, https://doi.org/10.1186/s12911-025-03072-x.
- R. Hu, Z. Deng, and X. Zhu, “Multi-scale graph fusion for co-saliency detection,” AAAI, vol. 35, no. 9, pp. 7789–7796, 2021, https://doi.org/10.1609/aaai.v35i9.16951.
- B. E. Dejene, T. M. Abuhay, and D. S. Bogale, “Predicting the level of anemia among Ethiopian pregnant women using homogeneous ensemble machine learning algorithm,” BMC Med. Inform. Decis. Mak., vol. 22, no. 1, p. 247, 2022, https://doi.org/10.1186/s12911-022-01992-6.
- S. Abraham and S. Joseph, “Medical Imaging and Artificial Intelligence: Transforming the Nature of Diagnostics and Treatment,” in Advances in Medical Technologies and Clinical Practice, pp. 127–158, 2024, https://doi.org/10.4018/979-8-3693-8990-4.ch006.
- S. Kukreti, A. Shrivastava, R. Chandrashekar, K. P. Rani, A. Badhoutiya, and S. Lakhanpal, “AI-Driven Clinical Decision Support Systems: Revolutionizing Healthcare With Predictive Models,” in 2025 International Conference on Computational, Communication and Information Technology (ICCCIT), pp. 560–565, 2025, https://doi.org/10.1109/ICCCIT62592.2025.10927929.
- Y. Yang, Y. Rao, M. Yu, and Y. Kang, “Multi-layer information fusion based on graph convolutional network for knowledge-driven herb recommendation,” Neural Netw., vol. 146, pp. 1–10, 2022, https://doi.org/10.1016/j.neunet.2021.11.010.
- H. Lin, K. Chen, Y. Xue, S. Zhong, L. Chen, and M. Ye, “Coronary heart disease prediction method fusing domain-adaptive transfer learning with graph convolutional networks (GCN),” Sci. Rep., vol. 13, no. 1, p. 14276, 2023, https://doi.org/10.1038/s41598-023-33124-z.
- Y. Huang and A. C. S. Chung, “Disease prediction with edge-variational graph convolutional networks,” Med. Image Anal., vol. 77, p. 102375, Apr. 2022, https://doi.org/10.1016/j.media.2022.102375.
- W. Hou, C. Lin, L. Yu, J. Qin, R. Yu, and L. Wang, “Hybrid Graph Convolutional Network With Online Masked Autoencoder for Robust Multimodal Cancer Survival Prediction,” IEEE Trans. Med. Imaging, vol. 42, no. 8, pp. 2462–2473, 2023, https://doi.org/10.1109/TMI.2023.3253760.
AUTHOR BIOGRAPHY

| Dimas Chaerul Ekty Saputra received his bachelor’s degree from the Department of Informatics, Faculty of Industrial Technology, Ahmad Dahlan University, Yogyakarta, Indonesia in 2020, an M.Sc. degree in Department of Biomedical Engineering, Graduate School, Universitas Gadjah Mada, Yogyakarta, Indonesia in 2022, and reveived his Ph.D in Computer Science and Information Technology, College of Computing, Khon Kaen University, Khon Kaen, Thailand in 2025. He is currently a lecturer at Telkom University, Surabaya Campus, Indonesia. He is also a member of the Association for Scientific Computing and Electronics, Engineering (ASCEE) Student Branch Indonesia. His research interests include artificial Intelligence, pattern recognition, machine learning, signal processing, and bioinformatics. Email: dimaschaerulekty@telkomuniversity.ac.id, Orcid: 0000-0001-6978-2846. |
|
|

| Irianna Futri received her bachelor’s degree in information systems from the Nurdin Hamzah School of Computer and Informatics Management in Jambi, Indonesia, in 2011. She worked at The Construction Services Public Works Department Jambi for 1 year as an Administrator for SIPJAKI (Construction Services Development Information System). She also worked as an informatics teacher at the Vocational School of Dharma Bhakti 4 for 3 years and served as an Operator for Dapodik for 7 years. She is pursuing a master’s degree in international technology and Innovation Management at the International College, Khon Kaen University, Khon Kaen, Thailand. Her current research interests include advanced care plans and bibliometric analysis. Email: irianna.f@kkumail.com, Orcid: 0009-0003-7100-3583. |
|
|

| Elvaro Islami Muryadi received his bachelor’s degree in public health, Respati Indonesia University, Jakarta, Indonesia, in 2015. Prior to that, in 2023, he completed a master's program in public health at Prima Indonesia University, Medan, Indonesia, after completing a master's program in business administration at Respati Indonesia University, Jakarta, Indonesia, in 2018. At present, he is engaged in doctoral studies at Khon Kaen University, Thailand, Faculty of Medicine, Department of Community, Occupational, and Family Medicine. Health education, public health, health promotion, and family medicine are among his current research interests. Email: elvaroislamimuryadi.e@kkumail.com, Orcid: 0000-0001-5554-3641. |
Dimas Chaerul Ekty Saputra (An Innovation Approach for Feature Selection Medical Data Using Joint Fine-Tuning Fusion Graph Convolutional Network)