Buletin Ilmiah Sarjana Teknik Elektro ISSN: 2685-9572
Comparative Analysis of Machine Learning Models for Tree Species Classification from UAV LiDAR Data
Gregorius Airlangga
Information System Study Program, Universitas Katolik Indonesia Atma Jaya, Indonesia
ARTICLE INFORMATION |
| ABSTRACT |
Article History: Submitted 13 January 2024 Revised 21 February 2024 Accepted 12 March 2024 |
| 
Forest ecosystems play a pivotal role in maintaining global biodiversity and climate balance. The precise identification of tree species via remote sensing technologies is vital for effective ecological surveillance and forest stewardship. This research conducts a comparative analysis of various machine learning algorithms for the binary classification of tree species utilizing LiDAR data captured by Unmanned Aerial Vehicles (UAVs). We analyzed a dataset featuring 192 trees from a diverse forest, employing models such as Logistic Regression, Support Vector Machine (SVM), Random Forest, K-Nearest Neighbors (KNN), Gradient Boosting, and Decision Trees. These models were assessed on their accuracy, precision, recall, and F1-scores to ascertain their efficacy. Our findings reveal that Logistic Regression and SVM were superior, achieving precision and recall scores up to 0.96, indicating their robust predictive capability. In contrast, KNN underperformed, suggesting the need for parameter refinement. Although ensemble methods demonstrated resilience, they were more prone to overfitting in comparison to the more straightforward Logistic Regression and SVM models. Preliminary data preprocessing and feature engineering techniques are discussed, enhancing the models' performance. This work enriches the domain of remote sensing and ecological monitoring by offering an in-depth evaluation of machine learning models for tree species classification, underscoring their advantages and constraints. It underscores the transformative potential of machine learning in refining ecological analysis precision, thereby aiding in the pursuit of sustainable forest management. Future research directions could include model refinement through advanced feature selection or the exploration of novel machine learning algorithms for improved classification accuracy. |
Keywords: UAV LiDAR Data; Tree Species Classification; Machine Learning; Remote Sensing; Forest Biodiversity |
Corresponding Author: Gregorius Airlangga, Universitas Katolik Indonesia Atma Jaya, Jakarta, Indonesia. Email: gregorius.airlangga@atmajaya.ac.id |
This work is licensed under a Creative Commons Attribution-Share Alike 4.0 
|
Document Citation: G. Airlangga, “Comparative Analysis of Machine Learning Models for Tree Species Classification from UAV LiDAR Data,” Buletin Ilmiah Sarjana Teknik Elektro, vol. 6, no. 1, pp. 54-62, 2024, DOI: 10.12928/biste.v6i1.10059. |
- INTRODUCTION
The essential role of forests in maintaining ecological balance, supporting biodiversity, and sequestering carbon has positioned forest conservation and management at the forefront of global environmental priorities [1]–[3]. Amidst the dual crises of climate change and biodiversity loss, the urgency to develop accurate, efficient, and scalable methods for monitoring forest health, structure, and species composition has intensified [4]–[6]. Advanced remote sensing technologies, particularly Light Detection and Ranging (LiDAR), offer unprecedented opportunities to address these challenges [7]–[9]. However, the full potential of LiDAR in discriminating between tree species in mixed and dense forests—a critical aspect for biodiversity assessments and ecological monitoring—remains underexploited [10]–[12]. This gap highlights the pressing need for innovative research that bridges advanced computational methods with ecological science to enhance our understanding and management of forest ecosystems [5],[13],[14]. The advent of high-resolution LiDAR technology has transformed ecological monitoring, allowing for detailed 3D representations of forest canopies [15]. Initial studies primarily focused on forest structure and biomass estimation, with species classification emerging as a pivotal area of interest more recently. The research from [15]–[17] have contributed foundational work on extracting and utilizing LiDAR-derived metrics for tree species identification, showcasing the potential of height distribution, canopy density, and textural features in distinguishing species. Despite these advancements, the application of these techniques to complex mixed forests, where species diversity and structural variability present significant challenges, has been less explored .
The integration of machine learning with LiDAR data for tree species classification has seen promising developments, with algorithms such as Random Forest, SVM, and KNN demonstrating varying degrees of success [18]. These studies underscore the importance of feature selection and the adaptability of models to complex datasets [19]. However, the literature reveals a fragmentation of approaches, with limited comprehensive comparisons across diverse forest types and species, highlighting a critical research gap in the field [20]. Machine learning models have increasingly become the cornerstone of remote sensing applications, offering robust tools for analyzing complex datasets. In the context of tree species classification, these models have the potential to learn from the intricate patterns within LiDAR data, such as the spatial arrangement of leaves and branches, which are indicative of specific species [21]. Ensemble methods, like Random Forest and Gradient Boosting, have been particularly noted for their ability to handle overfitting and improve prediction accuracy by aggregating results from multiple decision trees. Similarly, SVMs and KNNs offer powerful mechanisms for data classification, leveraging the geometric properties of the data space [22]–[24]. Yet, the effectiveness of these approaches can be significantly influenced by the quality of input features and the specific challenges presented by mixed forest environments, such as overlapping canopy layers and varied tree densities [25]. This points to a pressing need for a nuanced understanding of model performance in relation to forest complexity and species diversity [26].
While previous research has laid the groundwork for tree species classification using LiDAR data, a significant gap remains in the application and comparative analysis of machine learning models across mixed forest datasets [27]. The variability inherent in mixed forests, including species overlap, diverse canopy structures, and environmental noise, poses unique challenges that are yet to be fully addressed [28]. Moreover, the literature indicates a piecemeal approach to model testing, with studies often focusing on a single algorithm or a limited feature set. This research gap underscores the need for a holistic examination of how different machine learning models perform against the backdrop of a complex and biodiverse forest landscape, taking into account the full spectrum of LiDAR-derived features [29]. This study aims to systematically evaluate and compare the efficacy of various machine learning models for classifying tree species within a mixed forest environment, using UAV LiDAR data. By focusing on a comprehensive set of LiDAR-derived features and a dataset comprising individual trees from a dense mixed forest, the research seeks to identify optimal models and feature sets for accurate species classification. Through this endeavor, the study addresses a critical gap in the application of remote sensing technologies to forest biodiversity assessments, contributing to the development of more effective tools for ecological monitoring and management.
This research makes several key contributions to the field of ecological monitoring and forest management. Firstly, it introduces a uniquely curated dataset of individual trees from a UAV LiDAR survey of a dense mixed forest, providing a valuable resource for model training and validation. Secondly, it offers a comprehensive evaluation of multiple machine learning models, shedding light on their comparative effectiveness for tree species classification in mixed forests. These contributions not only advance our methodological approach to remote sensing in forestry but also enhance our capacity to conduct detailed biodiversity assessments, supporting conservation efforts and sustainable forest management practices. Following this introduction, the article is organized into several key sections: The Methodology section outlines the data collection process, feature extraction techniques, and the machine learning models employed. The Results section presents a detailed analysis of model performance, including classification accuracy and feature importance then, it also contextualizes the findings within the broader literature, exploring their implications for forest management and conservation. Finally, the Conclusion section summarizes the study's contributions and outlines avenues for further research, particularly in refining classification models and exploring their application to larger, more diverse forest datasets.
- METHODS
- Data Collection
In the realm of remote sensing and ecological analysis, the utilization of Unmanned Aerial Vehicle (UAV) Light Detection and Ranging (LiDAR) technology stands as a pioneering method for capturing intricate forest structures with high fidelity. The study harnessed this advanced technology to amass a comprehensive dataset, consisting of 192 individual trees meticulously distinguished from an extensive LiDAR point cloud. The data acquisition was performed using a state-of-the-art UAV platform equipped with a high-precision LiDAR sensor, specifically designed for ecological and forestry applications. The chosen UAV platform, coupled with the LiDAR sensor, enabled the collection of high-resolution point clouds, capturing the forest's structural complexity with exceptional detail. Point cloud data can be collected from [30]. This point cloud was acquired over a diverse, mixed forest landscape, encapsulating a rich variety of tree species in a dense forest setting. This dataset is distinguished by its diverse composition, featuring an almost equal distribution of coniferous and deciduous tree species. Each tree was identified and annotated with detailed species information, a process underpinned by thorough field surveys. Such detailed species-level annotation is crucial for the subsequent analysis, as it enables a targeted investigation into the structural differences between and within the species categories. The LiDAR technology facilitated the collection of detailed three-dimensional coordinates (X,Y,Z) for each point in the point cloud, alongside reflectance values that provide insights into the material characteristics of the forest canopy.
In terms of preprocessing, the point cloud data underwent several steps to ensure its suitability for analysis. These included the organization of data into a structured format, the classification of points belonging to individual trees, and the removal of irrelevant points, such as those not belonging to the vegetation layer. A key decision in this process was the establishment of a height threshold to exclude points from undergrowth and ground cover, enhancing the focus on the canopy and midstory vegetation. This threshold was determined based on field observations and preliminary analyses, which indicated that excluding points below a certain height would reduce noise and improve the classification accuracy of tree species. The three-dimensional coordinates captured by the LiDAR system enable researchers to reconstruct a precise digital representation of the forest's structural complexity. The X and Y coordinates represent the spatial positioning of each point within the horizontal plane, while the Z coordinate denotes the height of each point, offering a vertical dimension to the analysis. This three-dimensional data model is instrumental in assessing the canopy structure and density, which are critical factors in differentiating between tree species. Moreover, the reflectance values, which measure the intensity of light returned to the LiDAR sensor, add an additional layer of data that can be correlated with the physical characteristics of the trees.
- Preprocessing
The preprocessing of LiDAR data is a critical step in ensuring the accuracy and reliability of subsequent analyses. This phase entails a meticulous process designed to refine the raw data, making it more amenable to feature extraction and the application of machine learning models. The initial stage of preprocessing involves the systematic organization and sorting of the LiDAR files, which are typically stored in the .las file format. This organization is crucial for aligning the voluminous point cloud data with the corresponding tree species, a task facilitated by the directory structure that categorizes the data according to species identification. Such organization not only streamlines the analysis process but also enhances the efficiency of data retrieval and manipulation. Following the organization of the data, a critical step in the preprocessing workflow involves the classification of trees into two principal categories: coniferous and deciduous. This binary classification leverages a predefined list of species, enabling a focused approach to the analysis. The mathematical representation of this classification can be conceptualized as a function f: S→{0,1}, where S is the set of all species, and the function maps each species to either 0 (coniferous) or 1 (deciduous). This categorization forms the basis for binary-class classification tasks, simplifying the complex diversity of forest species into two manageable groups for analytical purposes.
An additional preprocessing step involves the removal of points that do not contribute meaningfully to the analysis of canopy structure. Specifically, points falling below a certain height threshold—typically set at one meter—are excluded from the dataset. This exclusion criterion is based on the rationale that points below this threshold are likely to represent ground noise or underbrush rather than meaningful components of the tree canopy. Mathematically, this filtering process can be described by the condition 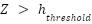 , where
, where 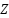 represents the height coordinate of each point in the point cloud, and
represents the height coordinate of each point in the point cloud, and 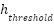 is the predefined height threshold. By applying this condition, the dataset is refined to include only those points that are relevant for analyzing the tree canopy, thereby enhancing the quality and relevance of the data for further analysis. These preprocessing steps collectively serve to enhance the integrity and utility of the LiDAR data, setting a solid foundation for the extraction of meaningful features and the application of sophisticated machine learning algorithms. By systematically organizing, classifying, and filtering the data, the study ensures that the subsequent analyses are based on accurate, relevant, and high-quality data, thereby increasing the likelihood of deriving insightful and reliable findings from the study.
is the predefined height threshold. By applying this condition, the dataset is refined to include only those points that are relevant for analyzing the tree canopy, thereby enhancing the quality and relevance of the data for further analysis. These preprocessing steps collectively serve to enhance the integrity and utility of the LiDAR data, setting a solid foundation for the extraction of meaningful features and the application of sophisticated machine learning algorithms. By systematically organizing, classifying, and filtering the data, the study ensures that the subsequent analyses are based on accurate, relevant, and high-quality data, thereby increasing the likelihood of deriving insightful and reliable findings from the study.
- Feature Extraction
A critical aspect of the study was the extraction of a comprehensive set of features from the cleaned LiDAR point clouds. These features were meticulously chosen to encapsulate the structural and spatial characteristics unique to each tree species. The extracted features included a range of height distribution metrics such as maximum height, mean, standard deviation, skewness, kurtosis, and entropy of the Z coordinates. Additional metrics such as the percentage of points above the mean height and specific height thresholds provided further insights into the vertical structure of the tree canopy. The study also derived eigenvalue features from the covariance matrix of the point clouds, offering a sophisticated analysis of the tree's shape and orientation through metrics like linearity, planarity, scatter, omnivariance, eigentropy, the sum of eigenvalues, and curvature. These features were selected for their demonstrated effectiveness in previous studies at distinguishing between tree species and capturing the complex architecture of forest canopies.
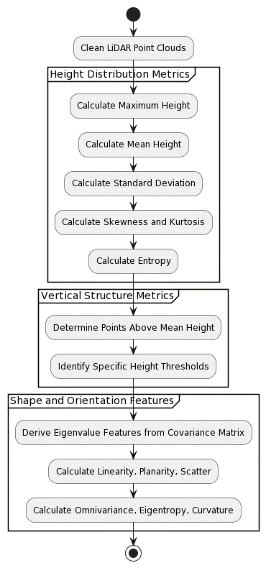
As presented in the Figure 1, the feature extraction process depicted in the image is a structured approach to analyzing LiDAR point cloud data, crucial for classifying tree species based on their unique structural and spatial characteristics.
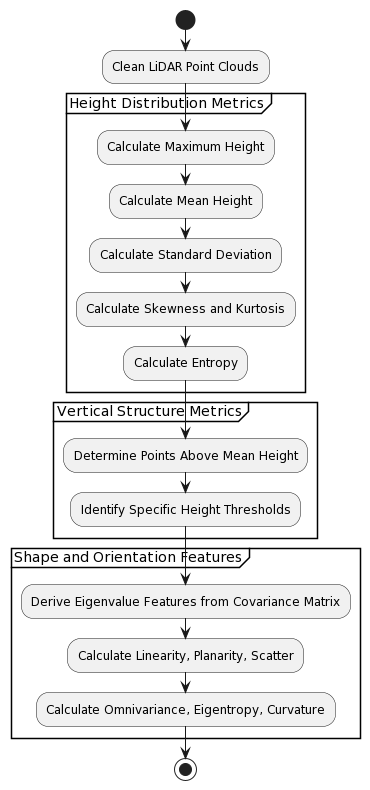
Figure 1. Feature Extraction Method
The flow begins with the cleaning of LiDAR point clouds to remove noise and irrelevant points, ensuring that the data used in analysis is of high quality and free of distortions that could affect the outcome. Upon cleaning, the process continues with the extraction of height distribution metrics. These metrics are essential as they provide a detailed understanding of the trees' vertical characteristics, which are critical for species identification. The maximum height of each tree is determined, representing the tallest point within the point cloud. The mean height is then computed to ascertain the average canopy height, while the standard deviation assesses the variability in height distribution across the canopy. Further statistical analysis includes calculating skewness and kurtosis, which give insights into the asymmetry and peakedness of the height distribution. Lastly, entropy is calculated to measure the randomness in height distribution, which can indicate the complexity of the tree's structure.
Following the extraction of height metrics, the focus shifts to vertical structure metrics. These metrics delve into the density and layering of the canopy by determining the percentage of points that are above the average height. Additionally, points are classified according to specific height thresholds, which helps to understand the stratification and layering within the canopy, an important aspect for distinguishing between species. The final phase of the feature extraction involves deriving shape and orientation features from the covariance matrix of the point cloud data. Sophisticated mathematical computations yield eigenvalue features that describe the tree's three-dimensional shape and orientation. Features such as linearity, planarity, and scatter are indicative of the tree's overall form, whether it is elongated, flat, or dispersed. Furthermore, omnivariance, eigentropy, and curvature provide a nuanced understanding of the tree's canopy complexity and the smoothness of its shape.
- Model Development and Evaluation
The study employed a diverse array of machine learning models to classify tree species based on the extracted LiDAR features. These models included Random Forest, Support Vector Machine (SVM) with a Linear Kernel, K-Nearest Neighbors (KNN), Gradient Boosting, and Decision Tree as presented in the equation (4) – equation (9). In the pursuit of accurately classifying tree species from LiDAR-derived features, our study curated a selection of machine learning models, each chosen for its proven record in handling classification tasks with high-dimensional data, such as that obtained from LiDAR. The ensemble method Random Forest was selected due to its robustness and ability to model non-linear relationships without extensive parameter tuning. It operates by constructing a multitude of decision trees at training time and outputting the class that is the mode of the classes of the individual trees, which inherently manages overfitting. The Support Vector Machine (SVM) with a Linear Kernel was included for its effectiveness in high-dimensional spaces and its ability to handle sparse data, which is typical in remote sensing applications. SVMs are particularly known for their capability to create optimal hyperplanes in a multidimensional space, which acts as a decision boundary between various classes. K-Nearest Neighbors (KNN) was employed for its simplicity and efficiency in classifying data based on feature similarity. This method is intuitive and non-parametric, making it suitable for classification where the decision boundary is not clearly defined. Gradient Boosting was chosen for its predictive power and ability to optimize on different loss functions. As a sequential ensemble method that corrects its predecessors' mistakes, Gradient Boosting builds strong predictive models, which is essential when dealing with diverse forest data. Lastly, the Decision Tree classifier was utilized for its interpretability and ease of use. As a model that uses a tree-like graph of decisions, it’s especially useful in understanding the feature space and the role of different features in species classification.
Each model was encapsulated within a pipeline that incorporated standard scaling of features to normalize the dataset as presented in the equation (1), ensuring that the classifiers could effectively process the varying scales of the LiDAR-derived features. The dataset was strategically split into training and test sets, with a 60-40 ratio as presented in the equation (2) – equation (3), ensuring that both sets were stratified by species to maintain an accurate representation of each species' proportion. The models were trained on the training set, and their performance was meticulously evaluated on the test set using a suite of metrics including accuracy, precision, recall, and F1 score as presented in the equation (10) – equation (13). This evaluation provided a detailed assessment of each model's effectiveness in accurately distinguishing between the different tree species, contributing to the study's aim of identifying the optimal machine learning approach for tree species classification in mixed forest environments.
| 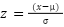
| (1) |
| 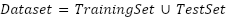
| (2) |
| 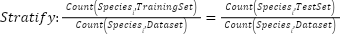
| (3) |
| 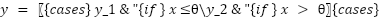
| (4) |
| 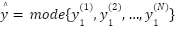
| (5) |
| 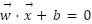
| (6) |
| 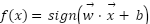
| (7) |
| 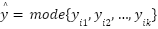
| (8) |
| 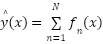
| (9) |
| 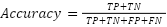
| (10) |
| 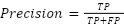
| (11) |
| 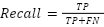
| (12) |
| 
| (13) |
- RESULT AND DISCUSSION
The results displayed in Table 1 provide a comparative analysis of various machine learning models used for binary classification tasks in the context of tree species identification from LiDAR data. The Table 1 outlines the performance of each model across four key metrics: accuracy, precision, recall, and F1-score. These metrics are essential for understanding how well each model is performing, with each metric providing insight into different aspects of the model's predictive abilities. Accuracy is the most intuitive performance measure and it is simply a ratio of correctly predicted observation to the total observations. It is suitable when the target classes are well balanced. The Logistic Regression and Support Vector Machine (SVM) models show the highest accuracy at 0.96, indicating that 96% of their predictions were correct. The Random Forest and Gradient Boosting models follow closely with 0.95 and 0.94 accuracy, respectively. The Decision Tree model has a slightly lower accuracy of 0.91, and the K-Nearest Neighbors (KNN) model has the lowest accuracy of 0.88, which could be indicative of overfitting or the model's sensitivity to the dataset's noise.
Furthermore, Precision is the ratio of correctly predicted positive observations to the total predicted positive observations. High precision relates to a low false positive rate, and the Logistic Regression, SVM, and Random Forest models lead with a precision of 0.96 and 0.95 respectively. This high precision indicates that when these models predict a tree species class, they are very likely to be correct. Gradient Boosting and Decision Tree models follow with a precision of 0.94 and 0.91, while the KNN model again trails at 0.89.Recall (also known as sensitivity) is the ratio of correctly predicted positive observations to all observations in actual class. It shows the model's ability to find all the positive samples. The Logistic Regression and SVM models demonstrate the highest recall of 0.96, suggesting that they are able to identify 96% of all actual positives in the dataset. Random Forest also performs well with a recall of 0.95. Gradient Boosting and Decision Tree models present a recall of 0.93 and 0.91, indicating slightly lower sensitivity, and KNN has the lowest with 0.89.The F1-score is the weighted average of Precision and Recall. Therefore, this score takes both false positives and false negatives into account. It is especially useful when the class distribution is uneven. The F1-score is the harmonic mean of precision and recall, with the best value at 1 and worst at 0. The Logistic Regression and SVM models achieve the highest F1-score at 0.96, which suggests a balanced performance between precision and recall. Random Forest shows a slightly lower F1-score of 0.95, followed by Gradient Boosting at 0.93, Decision Tree at 0.91, and KNN at 0.88.
Tabel 1. Comparison Result of Binary Classification
Methods | Accuracy | Precision | Recall | F1-Score |
Logistic Regression | 0.96 | 0.96 | 0.96 | 0.96 |
Random Forest | 0.95 | 0.95 | 0.95 | 0.95 |
SVM | 0.96 | 0.96 | 0.96 | 0.96 |
KNN | 0.88 | 0.89 | 0.89 | 0.88 |
Gradient Boosting | 0.94 | 0.94 | 0.93 | 0.93 |
Decision Tree | 0.91 | 0.91 | 0.91 | 0.91 |
The results suggest that Logistic Regression and SVM are the most effective models for this particular binary classification task, showing top performance across all metrics. This could be due to the linear nature of the decision boundary in the feature space of the dataset. The high precision and recall indicate that these models are not only accurate but also consistent in their predictions across different classes. Random Forest and Gradient Boosting also show strong performance, which may be attributed to their ensemble nature, allowing them to handle the variance in the dataset effectively. The slightly lower scores in comparison to Logistic Regression and SVM might be due to the complexity of the models, which can lead to overfitting, especially in a dataset with intricate structures such as LiDAR point clouds representing tree species. The Decision Tree model, while generally robust, shows lower performance compared to its ensemble counterparts. This may be because Decision Trees are prone to overfitting, particularly in the case of datasets with complex feature spaces. The results imply that the Decision Tree model may not capture the underlying patterns as effectively as the other models. The KNN model's lower performance could be due to the high dimensionality of the feature space. KNN is sensitive to the local structure of the data, and if the feature space is too large, the distance metric used to identify the 'nearest neighbors' becomes less meaningful. This can result in poorer performance on both precision and recall. The results emphasize the importance of model selection in machine learning tasks. While Logistic Regression and SVM models perform well in this scenario, it's crucial to note that model performance can vary significantly with changes in data distribution, feature engineering, and model hyperparameter tuning. Additionally, the dataset size, noise, and class imbalance can affect model performance.
- CONCLUSIONS
This study conducted a comparative analysis of several machine learning models to determine their efficacy in binary tree species classification using LiDAR data. Our findings reveal that Logistic Regression and SVM models outshine others in terms of precision and recall, indicating their robustness and suggesting their preferability for linearly separable datasets or those amenable to linear separation transformations. Conversely, models like KNN showed less impressive results, hinting at the need for refined feature selection and parameter tuning. Ensemble methods such as Random Forest and Gradient Boosting delivered solid performances; however, they fell short of the benchmarks set by Logistic Regression and SVM, likely due to their slight tendency towards overfitting and the complexity of hyperparameter optimization. The Decision Tree classifier's moderate success underscores the intricate nature of the dataset and its vulnerability to overfitting. By examining these models side-by-side, our research fills a critical gap in the literature on ecological monitoring and forest management, particularly in selecting suitable models for tree species classification. While the study illuminates the capabilities of various algorithms, it also acknowledges limitations. The performance of the models could be influenced by the dataset's unique composition of tree species from a mixed forest, which may affect the generalizability of our results to other forest types or ecological datasets. For future studies, we advocate for an exploration of feature engineering's impact and the integration of additional datasets, like multispectral imagery, to enhance model performance. The robustness of these models across diverse forest landscapes and under different data conditions, such as varying levels of noise or data sparsity, represents another avenue for research. Investigating more complex models, including deep learning architectures capable of discerning nonlinear patterns in high-dimensional data, could offer deeper insights. Understanding the balance between model complexity, interpretability, and performance remains an important challenge and a valuable direction for future work in ecological data analysis and decision-making processes.
REFERENCES
- A. Ameray, Y. Bergeron, O. Valeria, M. Montoro Girona, and X. Cavard, “Forest carbon management: A review of silvicultural practices and management strategies across boreal, temperate and tropical forests,” Curr. For. Reports, pp. 1–22, 2021, https://doi.org/10.1007/s40725-021-00151-w.
- P. Biber et al., “Forest biodiversity, carbon sequestration, and wood production: modeling synergies and trade-offs for ten forest landscapes across Europe,” Front. Ecol. Evol., vol. 8, p. 547696, 2020, https://doi.org/10.3389/fevo.2020.547696.
- A. Raj et al., “Forest Biodiversity Conservation and Restoration: Policies, Plan, and Approaches,” Ecorestoration Sustain., pp. 317–350, 2023, https://doi.org/10.1002/9781119879954.ch10.
- A. Wijerathna-Yapa and R. Pathirana, “Sustainable Agro-Food Systems for Addressing Climate Change and Food Security,” Agriculture, vol. 12, no. 10, p. 1554, 2022, https://doi.org/10.3390/agriculture12101554.
- S. E. Bibri, J. Krogstie, A. Kaboli, and A. Alahi, “Smarter eco-cities and their leading-edge artificial intelligence of things solutions for environmental sustainability: A comprehensive systematic review,” Environ. Sci. Ecotechnology, vol. 19, p. 100330, 2024, https://doi.org/10.1016/j.ese.2023.100330.
- S. E. Andres, et al., “Defining biodiverse reforestation: Why it matters for climate change mitigation and biodiversity,” Plants, People, Planet, vol. 5, no. 1, pp. 27-38, 2023, https://doi.org/10.1002/ppp3.10329.
- X. Liang et al., “Close-Range Remote Sensing of Forests: The state of the art, challenges, and opportunities for systems and data acquisitions,” IEEE Geosci. Remote Sens. Mag., vol. 10, no. 3, pp. 32–71, 2022, https://doi.org/10.1109/MGRS.2022.3168135.
- M. W. Rhodes, J. J. Bennie, A. Spalding, R. H. ffrench-Constant, and I. M. D. Maclean, “Recent advances in the remote sensing of insects,” Biol. Rev., vol. 97, no. 1, pp. 343–360, 2022, https://doi.org/10.1111/brv.12802.
- D. Xu, H. Wang, W. Xu, Z. Luan, and X. Xu, “LiDAR applications to estimate forest biomass at individual tree scale: Opportunities, challenges and future perspectives,” Forests, vol. 12, no. 5, p. 550, 2021, https://doi.org/10.3390/f12050550.
- M. Hirschmugl, C. Sobe, A. Di Filippo, V. Berger, H. Kirchmeir, and K. Vandekerkhove, “Review on the Possibilities of Mapping Old-Growth Temperate Forests by Remote Sensing in Europe,” Environmental Modeling & Assessment, vol. 28, no. 5, pp. 761-785, 2023, https://doi.org/10.1007/s10666-023-09897-y.
- K. Hwang et al., “Seeing the disturbed forest for the trees: Remote sensing is underutilized to quantify critical zone response to unprecedented disturbance,” Earth’s Futur., vol. 11, no. 8, p. e2022EF003314, 2023, https://doi.org/10.1029/2022EF003314.
- S. Ecke et al., “UAV-based forest health monitoring: A systematic review,” Remote Sens., vol. 14, no. 13, p. 3205, 2022, https://doi.org/10.3390/rs14133205.
- A. Shyrokaya et al., “Advances and gaps in the science and practice of impact-based forecasting of droughts,” Wiley Interdiscip. Rev. Water, p. e1698, 2023, https://doi.org/10.5194/ems2023-609.
- Y. K. Dwivedi et al., ““Real impact”: Challenges and opportunities in bridging the gap between research and practice–Making a difference in industry, policy, and society,” International Journal of Information Management, p. 102750, 2024, https://doi.org/10.1016/j.ijinfomgt.2023.102750.
- N. Camarretta et al., “Monitoring forest structure to guide adaptive management of forest restoration: a review of remote sensing approaches,” New Forests, vol. 51, no. 4, pp. 573-596, 2020, https://doi.org/10.1007/s11056-019-09754-5.
- C. T. de Almeida et al., “Combining LiDAR and hyperspectral data for aboveground biomass modeling in the Brazilian Amazon using different regression algorithms,” Remote Sensing of Environment, vol. 232, p. 111323, 2019, https://doi.org/10.1016/j.rse.2019.111323.
- Q. Li, B. Hu, J. Shang, and H. Li, “Fusion Approaches to Individual Tree Species Classification Using Multisource Remote Sensing Data,” Forests, vol. 14, no. 7, p. 1392, 2023, https://doi.org/10.3390/f14071392.
- K. K. McLauchlan et al., “ Fire as a fundamental ecological process: Research advances and frontiers,” Journal of Ecology, vol. 108, no. 5, pp. 2047–2069, 2020, https://doi.org/10.1111/1365-2745.13403.
- R. Pereira Martins-Neto, A. M. Garcia Tommaselli, N. N. Imai, E. Honkavaara, M. Miltiadou, E. A. Saito Moriya, and H. C. David, “Tree Species Classification in a Complex Brazilian Tropical Forest Using Hyperspectral and LiDAR Data,” Forests, vol. 14, no. 5, p. 945, 2023, https://doi.org/10.3390/f14050945.
- I. Tsamardinos et al., “Just Add Data: automated predictive modeling for knowledge discovery and feature selection,” NPJ Precis. Oncol., vol. 6, no. 1, p. 38, 2022, https://doi.org/10.1038/s41698-022-00274-8.
- Y. Shennan-Farpón, P. Visconti, and K. Norris, “Detecting ecological thresholds for biodiversity in tropical forests: Knowledge gaps and future directions,” Biotropica, vol. 53, no. 5, pp. 1276–1289, 2021, https://doi.org/10.1111/btp.12999.
- R. Hologa, K. Scheffczyk, C. Dreiser, and S. Gärtner, “Tree species classification in a temperate mixed mountain forest landscape using random forest and multiple datasets,” Remote Sens., vol. 13, no. 22, p. 4657, 2021, https://doi.org/10.3390/rs13224657.
- D. Xia, J. Shi, K. Wan, J. Wan, M. Martínez-García and X. Guan, "Digital Twin and Artificial Intelligence for Intelligent Planning and Energy-Efficient Deployment of 6G Networks in Smart Factories," in IEEE Wireless Communications, vol. 30, no. 3, pp. 171-179, 2023, https://doi.org/10.1109/MWC.017.2200495.
- I. Souiden, M. N. Omri, and Z. Brahmi, “A survey of outlier detection in high dimensional data streams,” Comput. Sci. Rev., vol. 44, p. 100463, 2022, https://doi.org/10.1016/j.cosrev.2022.100463.
- V. A. Surdu and R. Győrgy, “X-ray diffraction data analysis by machine learning methods—a review,” Appl. Sci., vol. 13, no. 17, p. 9992, 2023, https://doi.org/10.3390/app13179992.
- I. Fayad et al., “Hy-TeC: a hybrid vision transformer model for high-resolution and large-scale mapping of canopy height,” Remote Sens. Environ., vol. 302, p. 113945, 2024, https://doi.org/10.1016/j.rse.2023.113945.
- R. Seidl and M. G. Turner, “Post-disturbance reorganization of forest ecosystems in a changing world,” Proc. Natl. Acad. Sci., vol. 119, no. 28, p. e2202190119, 2022, https://doi.org/10.1073/pnas.2202190119.
- M. Immitzer and C. Atzberger, “Tree Species Diversity Mapping—Success Stories and Possible Ways Forward,” Remote Sens., vol. 15, no. 12, p. 3074, 2023, https://doi.org/10.3390/rs15123074.
- A. Shaamala, T. Yigitcanlar, A. Nili, and D. Nyandega, “Algorithmic Green Infrastructure Optimisation: Review of Artificial Intelligence Driven Approaches for Tackling Climate Change,” Sustain. Cities Soc., p. 105182, 2024, https://doi.org/10.1016/j.scs.2024.105182.
- Y. Hao, F. R. A. Widagdo, X. Liu, Y. Liu, L. Dong and F. Li, "A Hierarchical Region-Merging Algorithm for 3-D Segmentation of Individual Trees Using UAV-LiDAR Point Clouds," in IEEE Transactions on Geoscience and Remote Sensing, vol. 60, pp. 1-16, no. 5701416, 2022, https://doi.org/10.1109/TGRS.2021.3121419.
AUTHOR BIOGRAPHY

| GREGORIUS AIRLANGGA is received the B.S. degree in information system from the Yos Sudarso Higher School of Computer Science, Purwokerto, Indonesia, in 2014, and the M.Eng. degree in informatics from Atma Jaya Yogyakarta University, Yogyakarta, Indonesia, in 2016. He got Ph.D. degree with the Department of Electrical Engineering, National Chung Cheng University, Taiwan. He is also an Assistant Professor with the Department of Information System, Atma Jaya Catholic University of Indonesia, Jakarta, Indonesia. His research interests include data science, artificial intelligence and software engineering include path planning, machine learning, natural language processing, deep learning, software requirements, software design pattern and software architecture. |
Comparative Analysis of Machine Learning Models for Tree Species Classification from UAV LiDAR Data (Gregorius Airlangga)