Pipeline on microarray data analysis: Pre-processing
DOI:
https://doi.org/10.12928/bamme.v5i1.12539Keywords:
affymetrix, bioinformatics, microarray, pre-processingAbstract
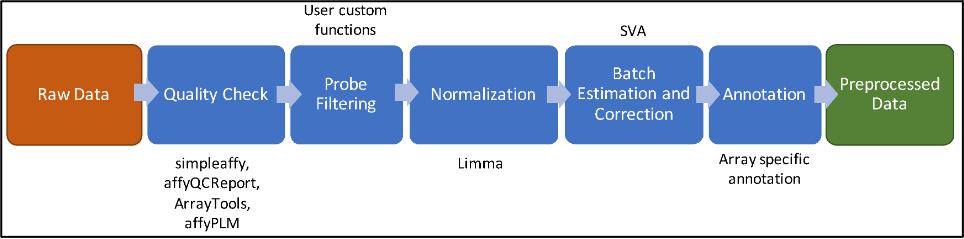
Bioinformatics is blooming and its data are store in some repository offline and or online. Yet some basic concepts are not fully disseminated. The paper intends to provide the reader with a review of one important concept in the pipeline bioinformatics data analysis of microarray, pre-processing. In pre-processing, there are four steps, background correction, normalization, probe correction and summarization. Each step consists of several methods, and we describe each method to give a better understanding on how it works theoretically. We focused on microarray data from Affymetrix platform with single-color chip.
References
Affymetrix, I. (2002). Statistical algorithms description document. Technical paper, 62, 110.
Astrand, M. (2003). Contrast normalization of oligonucleotide arrays. Journal of Computational Biology, 10(1), 95–102. https://doi.org/10.1089/106652703763255697
Baans, O. S., Jambek, A. B., & Said, K. A. M. (2019). Analysis of normalization method for DNA microarray data. Asia-Pacific Journal of Molecular Biology and Biotechnology, 27(4), 30–37. https://doi.org/10.35118/apjmbb.2019.027.4.04
Barbacioru, C. C., Wang, Y., Canales, R. D., Sun, Y. A., Keys, D. N., Chan, F., Poulter, K. A., & Samaha, R. R. (2006). Effect of various normalization methods on Applied Biosystems expression array system data. BMC Bioinformatics, 7, 1–14. https://doi.org/10.1186/1471-2105-7-533
Bolstad, B. M. (2004). Bolstad_2004_Dissertation. 156. papers2://publication/uuid/8B996D4A-CD91-4F11-9F50-7B5E60EFC00C
Bolstad, B. M., Irizarry, R. A., Astrand, M., & Speed, T. P. (2003). Gene Expression Omnibus A Comparison of Normalization Methods for High Density Oligonucleotide Array Data Based on Bias and Variance. Bioinformatics, 19(2), 185–193. http://www.ncbi.nlm.nih.gov/geo
Chen, Z., McGee, M., Liu, Q., & Scheuermann, R. H. (2007). A distribution free summarization method for Affymetrix GeneChip® arrays. Bioinformatics, 23(3), 321–327. https://doi.org/10.1093/bioinformatics/btl609
Cheng, L., Lo, L. Y., Tang, N. L. S., Wang, D., & Leung, K. S. (2016). CrossNorm: A novel normalization strategy for microarray data in cancers. Scientific Reports, 6, 1–2. https://doi.org/10.1038/srep18898
Cleveland, W. S. (1979). Robust Locally Weighted Regression and Smoothing Scatterplots. Journal of the American Statistical Association, 74(368), 829. https://doi.org/10.2307/2286407
Cleveland, W. S., & Devlin, S. J. (1988). Locally Weighted Regression: An Approach to Regression Analysis by Local Fitting. Journal of the American Statistical Association, 83(403), 596. https://doi.org/10.2307/2289282
Dozmorov, M. G., Guthridge, J. M., Hurst, R. E., & Dozmorov, I. M. (2010). A comprehensive and universal method for assessing the performance of differential gene expression analyses. PLoS ONE, 5(9), 1–11. https://doi.org/10.1371/journal.pone.0012657
Dudoit, S., Yang, Y. H., Speed, T. P., & Callow, M. J. (2002). Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Statistica Sinica, 12(1), 111–140. http://www1.cs.columbia.edu/~cleslie/cs4761/lectures/speed-statistical.pdf
Durbin, B. P., Hardin, J. S., Hawkins, D. M., & Rocke, D. M. (2002). A variance-stabilizing transformation for gene-expression microarray data. Bioinformatics (Oxford, England), 18 Suppl 1, S105–S110. https://doi.org/10.1093/bioinformatics/18.suppl_1.s105.
Fajriyah R. (2021). Paper review: An overview on microarray technologies. Bulletin of Applied Mathematics and Mathematics Education, 1(1), 21-30.
Federico, A., Saarimäki, L. A., Serra, A., Giudice, G. Del, Kinaret, P. A. S., Scala, G., & Greco, D. (2022). Microarray Data Preprocessing: From Experimental Design to Differential Analysis. 24(01), 79–100. https://doi.org/10.1007/978-1-0716-1839-4_7
Fujita, A., Sato, J. R., de Oliveira Rodrigues, L., Ferreira, C. E., & Sogayar, M. C. (2006). Evaluating different methods of microarray data normalization. BMC Bioinformatics, 7, 1–11. https://doi.org/10.1186/1471-2105-7-469
Gautier, L., Bolstad, B. M., Cope, L., & Irizarry, R. A. (2004). Affy - Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics, 20(3), 307–315. https://doi.org/10.1093/bioinformatics/btg405
Gharaibeh, R. Z., Fodor, A. A., & Gibas, C. J. (2008). Background correction using dinucleotide affinities improves the performance of GCRMA. BMC Bioinformatics, 9, 1–12. https://doi.org/10.1186/1471-2105-9-452
Giorgi, F.M., Bolger, A.M., Lohse, M. (2010). Algorithm-driven Artifacts in median polish summarization of Microarray data. BMC Bioinformatics, 11, 553. https://doi.org/10.1186/1471-2105-11-553.
Gondro, C. (2009). Summarization methods and quality problems in Affymetrix microarrays. Proc Assoc Advmt Anim Breed Genet, 18(February).
Grant, G. R., Manduchi, E., & Stoeckert, C. J. (2007). Analysis and management of microarray gene expression data. Current Protocols in Molecular Biology / Edited by Frederick M. Ausubel ... [et Al.], Chapter 19, 1–30. https://doi.org/10.1002/0471142727.mb1906s77
Hartemink, A. J., Gifford, D. K., Jaakkola, T. S., & Young, R. A. (2001). Maximum-likelihood estimation of optimal scaling factors for expression array normalization. Microarrays: Optical Technologies and Informatics, 4266(Ml), 132–140. https://doi.org/10.1117/12.427981
Hochreiter, S., Clevert, D. A., & Obermayer, K. (2006). A new summarization method for affymetrix probe level data. Bioinformatics, 22(8), 943–949. https://doi.org/10.1093/bioinformatics/btl033
Huber, W., Von Heydebreck, A., Sültmann, H., Poustka, A., & Vingron, M. (2002). Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics, 18(SUPPL. 1). https://doi.org/10.1093/bioinformatics/18.suppl_1.S96
Irizarry, R. A., Bolstad, B., Collin, F., Cope, L. M., Hobbs, B., & Speed, T. P. (2003). Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Research, 31(4), e15. https://doi.org/10.1093/nar/gng015
Klaus, B., & Reisenauer, S. (2018). An end to end workflow for differential gene expression using Affymetrix microarrays. F1000Research, 5, 1–56. https://doi.org/10.12688/f1000research.8967.2
Kuyuk, S. A. (2017). Commonly used statistical methods for detecting differential gene expression in microarray experiments. Biostatistics and Epidemiology International Journal, 0(0), 1–8. https://doi.org/10.30881/beij.00001
Li, C. and Wong, W. . (2001a). Model-based analysis of oligo- nucleotide arrays: expression index computation and outlier detection. Computational Statistics & Data Analysis, a(98), 31–36.
Li, C. and Wong, W. H. (2001b). Model-based analysis of oligo- nucleotide arrays: model validation, design issues and standard error application. b(2), 1-11.
Microarray Galaxy User’s Guide. (2023). Microarray Galaxy User’s Guide. http://www.ensat.ac.ma/mobihic/microarray-galaxy.html
Miranda, J., & Bringas, R. (2008). Analysis of DNA microarray data. Part I: Technological background and experimental design. Biotecnologia Aplicada, 25(2).
Munster, S., VL, W., Hutchings, DC., B. D., & Nicholson, S. (2018). Comparison Study of Microarray and RNA-seq for Differential Expression. Final Report. https://doi.org/DOT/FAA/AM-20/09
Wright Muelas, M., Mughal, F., O’Hagan, S. et al. The role and robustness of the Gini coefficient as an unbiased tool for the selection of Gini genes for normalising expression profiling data. Sci Rep 9, 17960 (2019). https://doi.org/10.1038/s41598-019-54288-7
Naef, F., & Magnasco, M. O. (2003). Solving the riddle of the bright mismatches: Labeling and effective binding in oligonucleotide arrays. Physical Review E - Statistical Physics, Plasmas, Fluids, and Related Interdisciplinary Topics, 68(1), 4. https://doi.org/10.1103/PhysRevE.68.011906
Olson, N. E. (2006). The Microarray Data Analysis Process: From Raw Data to Biological Significance. NeuroRx, 3(3), 373–383. https://doi.org/10.1016/j.nurx.2006.05.005
Pelz, C. R., Kulesz-Martin, M., Bagby, G., & Sears, R. C. (2008). Global rank-invariant set normalization (GRSN) to reduce systematic distortions in microarray data. BMC Bioinformatics, 9(January 2009). https://doi.org/10.1186/1471-2105-9-520
Piccolo, S. R., Ying Sun, Campbell, D, J., Lenburg, M. E., Bild, A. H., & W Evan Johnson. (2012). A single-sample microarray normalization method to facilitate personalized-medicine workflow. Genomics, 100(6), 337–344. https://doi.org/10.1016/j.ygeno.2012.08.003
Ritchie, M. E., Silver, J., Oshlack, A., Holmes, M., Diyagama, D., Holloway, A., & Smyth, G. K. (2007). A comparison of background correction methods for two-colour microarrays. Bioinformatics, 23(20), 2700–2707. https://doi.org/10.1093/bioinformatics/btm412
Serin, A. (2011). Biclustering Analysis for Large Scale Data. September. http://www.diss.fu-berlin.de/diss/receive/FUDISS_thesis_000000035625?lang=en
Silver JD, Ritchie ME, & S. G. (2009). Microarray background correction: maximum likelihood estimation for the normal-exponential convolution. Biostatistics and Epidemiology International Journal, 10(2), 52–63. https://doi.org/10.1093/biostatistics/kxn042
Smyth, G. K. (2006). Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray. 3(1), 1–26.
Smyth, G. K., & Speed, T. (2003). Normalization of cDNA microarray data. Methods, 31(4), 265–273. https://doi.org/10.1016/S1046-2023(03)00155-5
TechMedBuddy,. (2023). Microarray Data Analysis in Bioinformatics: A Comprehensive Overview. https://www.linkedin.com/pulse/microarray-data-analysis-overview-techmedbuddy/
Visentin, L., Scarpellino, G., Chinigò, G., Munaron, L., & Ruffinatti, F. A. (2022). BioTEA: Containerized Methods of Analysis for Microarray-Based Transcriptomics Data. Biology, 11(9), 1–14. https://doi.org/10.3390/biology11091346
Wright Muelas, M., Mughal, F., O’Hagan, S., Day, P. J., & Kell, D. B. (2019). The role and robustness of the Gini coefficient as an unbiased tool for the selection of Gini genes for normalising expression profiling data. Scientific Reports, 9(1), 1–21. https://doi.org/10.1038/s41598-019-54288-7
Wu, Z. (2009). A Review of Statistical Methods for Preprocessing. Nih, 71(2), 233–236. https://doi.org/10.1177/0962280209351924.A
Wu, Z., Irizarry, R. A., Gentleman, R., Martinez-Murillo, F., & Spencer, F. (2004). A model-based background adjustment for oligonucleotide expression arrays. Journal of the American Statistical Association, 99(468), 909–917. https://doi.org/10.1198/016214504000000683
Yang, J., & Thorne, N. (2002). Normalization for Two-color cDNA Microarray Data. Science and Statistics: A Festschrift for Terry Speed, 403–418.
Yang, Y., S, D., P, L., DM, L., V, P., J, N., & TP, S. (2002). Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Research, 30(4).
Downloads
Published
Issue
Section
License
Copyright (c) 2025 Rohmatul Fajriyah, Noodchanath Kongchouy, Wanvisa Saisanan Na Ayudhaya, Rahmadi Yotenka, Ghiffari Ahnaf Danarwindu

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
Authors who publish with this journal agree to the following terms:
- Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under Creative Commons Attribution License that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgement of its initial publication in this journal.
- Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See The Effect of Open Access).



